4BBJ
 
 | | Copper-transporting PIB-ATPase in complex with beryllium fluoride representing the E2P state | | Descriptor: | COPPER EFFLUX ATPASE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Mattle, D, Gourdon, P, Nissen, P. | | Deposit date: | 2012-09-25 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Copper-Transporting P-Type Atpases Use a Unique Ion-Release Pathway
Nat.Struct.Mol.Biol., 21, 2014
|
|
3USL
 
 | | Crystal Structure of LeuT bound to L-selenomethionine in space group C2 from lipid bicelles | | Descriptor: | ACETATE ION, IODIDE ION, PHOSPHOCHOLINE, ... | | Authors: | Wang, H, Elferich, J, Gouaux, E. | | Deposit date: | 2011-11-23 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UJ9
 
 | | Phosphoethanolamine methyltransferase from Plasmodium falciparum in complex with phosphocholine | | Descriptor: | PHOSPHOCHOLINE, Phosphoethanolamine N-methyltransferase | | Authors: | Lee, S.G, Kim, Y, Alpert, T.D, Nagata, A, Jez, J.M. | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target.
J.Biol.Chem., 287, 2012
|
|
3UJC
 
 | | Phosphoethanolamine methyltransferase mutant (H132A) from Plasmodium falciparum in complex with phosphocholine | | Descriptor: | PHOSPHOCHOLINE, Phosphoethanolamine N-methyltransferase | | Authors: | Lee, S.G, Kim, Y, Alpert, T.D, Nagata, A, Jez, J.M. | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target.
J.Biol.Chem., 287, 2012
|
|
3UJD
 
 | | Phosphoethanolamine methyltransferase mutant (Y19F) from Plasmodium falciparum in complex with phosphocholine | | Descriptor: | PHOSPHOCHOLINE, Phosphoethanolamine N-methyltransferase | | Authors: | Lee, S.G, Kim, Y, Alpert, T.D, Nagata, A, Jez, J.M. | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target.
J.Biol.Chem., 287, 2012
|
|
3USM
 
 | | Crystal Structure of LeuT bound to L-selenomethionine in space group C2 from lipid bicelles (collected at 1.2 A) | | Descriptor: | IODIDE ION, PHOSPHOCHOLINE, SELENOMETHIONINE, ... | | Authors: | Wang, H, Elferich, J, Gouaux, E. | | Deposit date: | 2011-11-23 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.008 Å) | | Cite: | Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2FF6
 
 | | Crystal structure of Gelsolin domain 1:ciboulot domain 2 hybrid in complex with actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Aguda, A.H, Xue, B, Robinson, R.C. | | Deposit date: | 2005-12-19 | | Release date: | 2006-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structural Basis of Actin Interaction with Multiple WH2/beta-Thymosin Motif-Containing Proteins
Structure, 14, 2006
|
|
3USG
 
 | | Crystal structure of LeuT bound to L-leucine in space group C2 from lipid bicelles | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, LEUCINE, ... | | Authors: | Wang, H, Elferich, J, Gouaux, E. | | Deposit date: | 2011-11-23 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2HG9
 
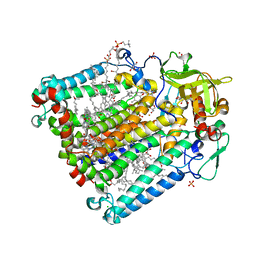 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with tetrabrominated phosphatidylcholine | | Descriptor: | (7R,18S,19R)-18,19-DIBROMO-7-{[(9S,10S)-9,10-DIBROMOOCTADECANOYL]OXY}-4-HYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-P HOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-26 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
4XCF
 
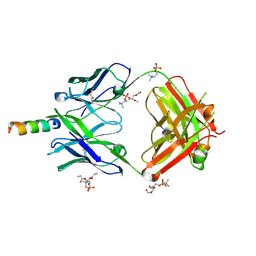 | | Crystal structure of human 4E10 Fab in complex with its peptide epitope on HIV-1 gp41; crystals cryoprotected with phosphatidylcholine (03:0 PC) | | Descriptor: | 4E10 Fab heavy chain, 4E10 Fab light chain, MODIFIED FRAGMENT OF HIV-1 GLYCOPROTEIN (GP41) INCLUDING THE MPER REGION 671-683, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-17 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
4UU0
 
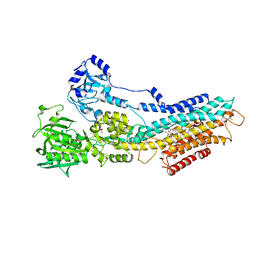 | | CRYSTAL STRUCTURE OF (SR) CALCIUM-ATPASE E2(TG) IN THE PRESENCE OF 14:1 PC | | Descriptor: | GLYCEROL, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Drachmann, N.D, Olesen, C, Moeller, J.V, Guo, Z, Nissen, P, Bublitz, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparing Crystal Structures of Ca(2+) -ATPase in the Presence of Different Lipids.
FEBS J., 281, 2014
|
|
5F3Z
 
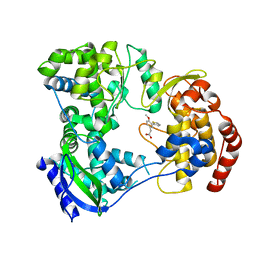 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to PC-79-SH52 | | Descriptor: | 2-(4-methoxy-3-thiophen-2-yl-phenyl)ethanoic acid, Genome polyprotein, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2015-12-03 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Conserved Pocket in the Dengue Virus Polymerase Identified through Fragment-based Screening
J.Biol.Chem., 291, 2016
|
|
5EZM
 
 | | Crystal Structure of ArnT from Cupriavidus metallidurans in the apo state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferases of PMT family, CHLORIDE ION, ... | | Authors: | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-26 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
5FFR
 
 | | Crystal Structure of Surfactant Protein-A complexed with phosphocholine | | Descriptor: | CALCIUM ION, PHOSPHOCHOLINE, Pulmonary surfactant-associated protein A, ... | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
7RZE
 
 | | Insulin Degrading Enzyme pO/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZF
 
 | | Insulin Degrading Enzyme O/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
6C74
 
 | |
7T7U
 
 | | Light Harvesting complex phycocyanin PC 630, from the cryptophyte Chroomonas sp. M1627 | | Descriptor: | DiCys-(15,16)-Dihydrobiliverdin, GLYCEROL, PHYCOCYANOBILIN, ... | | Authors: | Michie, K.A, Harrop, S.J, Rathbone, H.W, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2021-12-15 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular structures reveal the origin of spectral variation in cryptophyte light harvesting antenna proteins.
Protein Sci., 32, 2023
|
|
4TSP
 
 | | Crystal structure of FraC with DHPC bound (crystal form II) | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Fragaceatoxin C, PHOSPHATE ION, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
8VC1
 
 | | CryoEM structure of insect gustatory receptor BmGr9 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, ... | | Authors: | Frank, H.M, Walsh Jr, R.M, Garrity, P.A, Gaudet, R. | | Deposit date: | 2023-12-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of ligand specificity and channel activation in an insect gustatory receptor.
Cell Rep, 43, 2024
|
|
8WCQ
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in intermediate state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-09-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
6MIH
 
 | | Crystal structure of host-guest complex with PC hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*(1WA)P*CP*(DB)P*T)-3'), DNA (5'-D(P*AP*(DS)P*GP*(1W5)P*TP*AP*AP*G)-3'), N-terminal fragment of MMLV reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
6UE3
 
 | |
6VYM
 
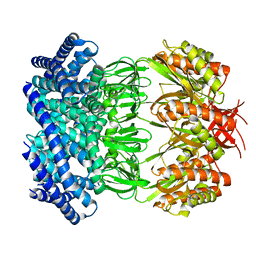 | | Cryo-EM structure of mechanosensitive channel MscS in PC-18:1 nanodiscs treated with beta-cyclodextran | | Descriptor: | Mechanosensitive channel MscS | | Authors: | Zhang, Y, Daday, C, Gu, R, Cox, C.D, Martinac, B, Groot, B, Walz, T. | | Deposit date: | 2020-02-27 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Visualization of the mechanosensitive ion channel MscS under membrane tension.
Nature, 590, 2021
|
|
6VYL
 
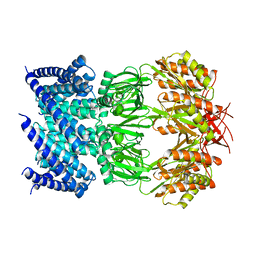 | | Cryo-EM structure of mechanosensitive channel MscS in PC-10 nanodiscs | | Descriptor: | Mechanosensitive channel MscS | | Authors: | Zhang, Y, Daday, C, Gu, R, Cox, C.D, Martinac, B, Groot, B, Walz, T. | | Deposit date: | 2020-02-27 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Visualization of the mechanosensitive ion channel MscS under membrane tension.
Nature, 590, 2021
|
|
