7ZC6
 
 | | Na+ - translocating ferredoxin: NAD+ reductase (Rnf) of C. tetanomorphum | | Descriptor: | FE (III) ION, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Ermler, U, Vitt, S, Buckel, W. | | Deposit date: | 2022-03-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Purification and structural characterization of the Na + -translocating ferredoxin: NAD + reductase (Rnf) complex of Clostridium tetanomorphum.
Nat Commun, 13, 2022
|
|
6RQR
 
 | | Extended NHERF1 PDZ2 domain in complex with the PDZ-binding motif of CFTR | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1,Cystic fibrosis transmembrane conductance regulator | | Authors: | Martin, E.R, Ford, R.C, Robinson, R.C. | | Deposit date: | 2019-05-16 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In vivocrystals reveal critical features of the interaction between cystic fibrosis transmembrane conductance regulator (CFTR) and the PDZ2 domain of Na+/H+exchange cofactor NHERF1.
J.Biol.Chem., 295, 2020
|
|
3EAD
 
 | | Crystal structure of CALX-CBD1 | | Descriptor: | CALCIUM ION, GLYCEROL, Na/Ca exchange protein | | Authors: | Zheng, L, Wang, M. | | Deposit date: | 2008-08-25 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of progressive Ca2+ binding states of the Ca2+ sensor Ca2+ binding domain 1 (CBD1) from the CALX Na+/Ca2+ exchanger reveal incremental conformational transitions.
J.Biol.Chem., 285, 2010
|
|
3E9T
 
 | | Crystal structure of Apo-form Calx CBD1 domain | | Descriptor: | CALCIUM ION, Na/Ca exchange protein | | Authors: | Wu, M, Zheng, L. | | Deposit date: | 2008-08-23 | | Release date: | 2009-09-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of progressive Ca2+ binding states of the Ca2+ sensor Ca2+ binding domain 1 (CBD1) from the CALX Na+/Ca2+ exchanger reveal incremental conformational transitions.
J.Biol.Chem., 285, 2010
|
|
1SFQ
 
 | | Fast form of thrombin mutant R(77a)A bound to PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, ... | | Authors: | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-02-20 | | Release date: | 2004-06-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
1SG8
 
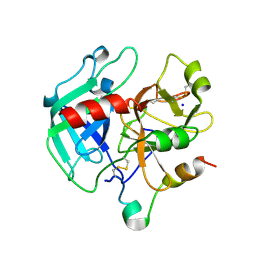 | | Crystal structure of the procoagulant fast form of thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, thrombin | | Authors: | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-02-23 | | Release date: | 2004-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
2E30
 
 | |
5JXE
 
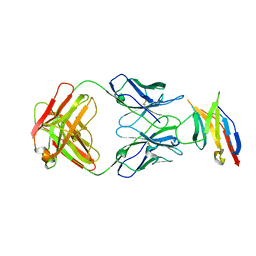 | | Human PD-1 ectodomain complexed with Pembrolizumab Fab | | Descriptor: | Pembrolizumab Fab heavy chain, Pembrolizumab Fab light chain, Programmed cell death protein 1 | | Authors: | Na, Z, Bharath, S.R, Song, H. | | Deposit date: | 2016-05-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for blocking PD-1-mediated immune suppression by therapeutic antibody pembrolizumab.
Cell Res., 27, 2017
|
|
6KNS
 
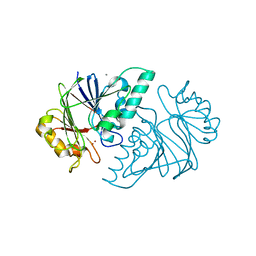 | | Crystal structure of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis (space group I4122) | | Descriptor: | CALCIUM ION, Putative metal-dependent hydrolase, ZINC ION | | Authors: | Na, H.W, Namgung, B, Song, W.S, Yoon, S.I. | | Deposit date: | 2019-08-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and biochemical analyses of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
5JOC
 
 | | Crystal structure of the S61A mutant of AmpC BER | | Descriptor: | Beta-lactamase, CITRIC ACID | | Authors: | Na, J.H, An, Y.J, Cha, S.S. | | Deposit date: | 2016-05-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the extended substrate spectrum of AmpC BER and structure-guided discovery of the inhibition activity of citrate against the class C beta-lactamases AmpC BER and CMY-10.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6KNT
 
 | | Crystal structure of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis (space group P4332) | | Descriptor: | Putative metal-dependent hydrolase, ZINC ION | | Authors: | Na, H.W, Namgung, B, Song, W.S, Yoon, S.I. | | Deposit date: | 2019-08-07 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
5WU7
 
 | |
4US3
 
 | | Crystal Structure of the bacterial NSS member MhsT in an Occluded Inward-Facing State | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, SODIUM ION, TRANSPORTER, ... | | Authors: | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
3ZK2
 
 | |
7X23
 
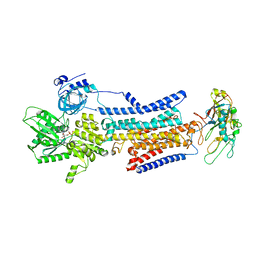 | | Cryo-EM structure of non gastric H,K-ATPase alpha2 SPWC mutant in 3Na+E1-AMPPCPF state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Potassium-transporting ATPase alpha chain 2, ... | | Authors: | Abe, K, Nakanishi, H, Young, V, Artigas, P. | | Deposit date: | 2022-02-25 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and function of H + /K + pump mutants reveal Na + /K + pump mechanisms.
Nat Commun, 13, 2022
|
|
2KBV
 
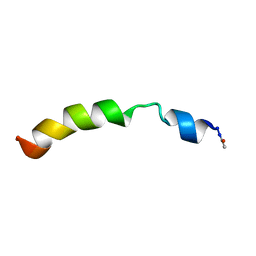 | | Structural and functional analysis of TM XI of the NHE1 isoform of thE NA+/H+ exchanger | | Descriptor: | Sodium/hydrogen exchanger 1 | | Authors: | Lee, B.L, Li, X, Liu, Y, Sykes, B.D, Fliegel, L. | | Deposit date: | 2008-12-09 | | Release date: | 2009-01-27 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Analysis of Transmembrane XI of the NHE1 Isoform of the Na+/H+ Exchanger
J.Biol.Chem., 284, 2009
|
|
2K3C
 
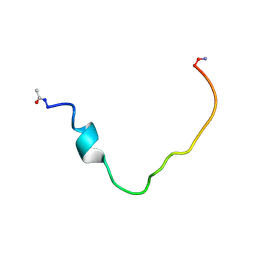 | | Structural and Functional Characterization of TM IX of the NHE1 Isoform of the Na+/H+ Exchanger | | Descriptor: | TMIX peptide | | Authors: | Reddy, T, Ding, J, Li, X, Sykes, B.D, Fliegel, L, Rainey, J.K. | | Deposit date: | 2008-05-01 | | Release date: | 2008-06-03 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of Transmembrane Segment IX of the NHE1 Isoform of the Na+/H+ Exchanger.
J.Biol.Chem., 283, 2008
|
|
8IWO
 
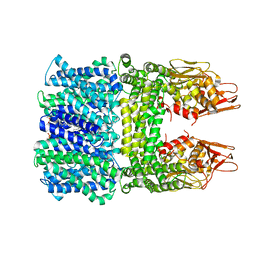 | | The rice Na+/H+ antiporter SOS1 in an auto-inhibited state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, OsSOS1 | | Authors: | Zhang, X.Y, Tang, L.H, Zhang, C.R, Nie, J.W, Chen, Y.H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structure and activation mechanism of the rice Salt Overly Sensitive 1 (SOS1) Na + /H + antiporter.
Nat.Plants, 9, 2023
|
|
3F6U
 
 | | Crystal structure of human Activated Protein C (APC) complexed with PPACK | | Descriptor: | CALCIUM ION, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, ... | | Authors: | Schmidt, A.E, Padmanabhan, K, Underwood, M.C, Bode, W, Mather, T, Bajaj, S.P. | | Deposit date: | 2008-11-06 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thermodynamic linkage between the S1 site, the Na+ site, and the Ca2+ site in the protease domain of human activated protein C (APC).
J.Biol.Chem., 277, 2002
|
|
2JO1
 
 | | Structure of the Na,K-ATPase regulatory protein FXYD1 in micelles | | Descriptor: | Phospholemman | | Authors: | Teriete, P, Franzin, C.M, Choi, J, Marassi, F.M. | | Deposit date: | 2007-02-18 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the Na,K-ATPase regulatory protein FXYD1 in micelles
Biochemistry, 46, 2007
|
|
4US4
 
 | | Crystal Structure of the Bacterial NSS Member MhsT in an Occluded Inward-Facing State (lipidic cubic phase form) | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, SODIUM ION, ... | | Authors: | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
9LRR
 
 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase NqrB-G141A mutant from Vibrio cholerae with bound korormicin A | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-02-01 | | Release date: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural elucidation of the mechanism for inhibitor resistance in the Na+-translocating NADH-ubiquinone oxidoreductase from Vibrio cholerae
To Be Published
|
|
8A1X
 
 | | Sodium pumping NADH-quinone oxidoreductase with inhibitor DQA | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1Y
 
 | | Sodium pumping NADH-quinone oxidoreductase with inhibitor HQNO | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4XCP
 
 | | Fatty Acid and Retinol binding protein Na-FAR-1 from Necator americanus | | Descriptor: | Nematode fatty acid retinoid binding protein, PALMITIC ACID | | Authors: | Gabrielsen, M, Rey-Burusco, M.F, Ibanez-Shimabukuro, M, Griffiths, K, Kennedy, M.W, Corsico, B, Smith, B.O. | | Deposit date: | 2014-12-18 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Diversity in the structures and ligand-binding sites of nematode fatty acid and retinol-binding proteins revealed by Na-FAR-1 from Necator americanus.
Biochem.J., 471, 2015
|
|
