3TUB
 
 | |
2WZO
 
 | | The structure of the FYR domain | | Descriptor: | GLYCEROL, TRANSFORMING GROWTH FACTOR BETA REGULATOR 1 | | Authors: | Garcia-Alai, M.M, Allen, M.D, Joerger, A.C, Bycroft, M. | | Deposit date: | 2009-12-01 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of the Fyr Domain of Transforming Growth Factor Beta Regulator 1 (Tbrg1)(Pna)
Protein Sci., 19, 2010
|
|
4CMO
 
 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor 2-((1R)-1-((3-amino-6-(2-methoxypyridin-3-yl)pyrazin-2-yl) oxy)ethyl)-4-fluoro-N-methylbenzamide | | Descriptor: | 2-[(1R)-1-{[3-amino-6-(2-methoxypyridin-3-yl)pyrazin-2-yl]oxy}ethyl]-4-fluoro-N-methylbenzamide, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-01-16 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
1HOW
 
 | | THE X-RAY CRYSTAL STRUCTURE OF SKY1P, AN SR PROTEIN KINASE IN YEAST | | Descriptor: | 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE YMR216C, SULFATE ION | | Authors: | Nolen, B.J, Yun, C.Y, Wong, C.F, McCammon, J.A, Fu, X.-D, Ghosh, G. | | Deposit date: | 2000-12-11 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of Sky1p reveals a novel mechanism for constitutive activity.
Nat.Struct.Biol., 8, 2001
|
|
1NAI
 
 | | UDP-GALACTOSE 4-EPIMERASE FROM ESCHERICHIA COLI, OXIDIZED | | Descriptor: | 1,3-PROPANDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Frey, P.A, Holden, H.M. | | Deposit date: | 1995-11-22 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the oxidized and reduced forms of UDP-galactose 4-epimerase isolated from Escherichia coli.
Biochemistry, 35, 1996
|
|
2G47
 
 | |
6JOQ
 
 | | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+ complex provide molecular mechanisms for substrate specificity | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Primase | | Authors: | Guo, H.J, Li, M.J, Wu, H, Yu, F, He, J.H. | | Deposit date: | 2019-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+complex provide molecular mechanisms for substrate specificity.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
5ZOF
 
 | | Crystal Structure of D181A/R192F hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*G)-3'), DNA (5'-D(*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), DNA (5'-D(*GP*CP*CP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
7ZMB
 
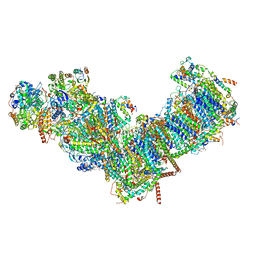 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZM7
 
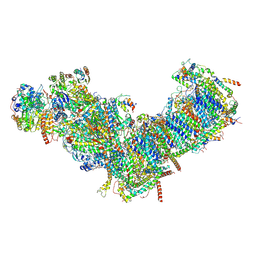 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
2QJI
 
 | | M. jannaschii ADH synthase complexed with dihydroxyacetone phosphate and glycerol | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, GLYCEROL, Putative aldolase MJ0400 | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2007-07-07 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonic acid synthase, a catalyst in the archaeal pathway for the biosynthesis of aromatic amino acids.
Biochemistry, 46, 2007
|
|
3BUF
 
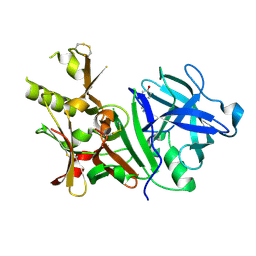 | | BACE-1 complexed with compound 2 | | Descriptor: | 4-[(2R)-2-aminopropyl]phenol, Beta-secretase 1 | | Authors: | Kuglstatter, A, Hennig, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tyramine fragment binding to BACE-1
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7LER
 
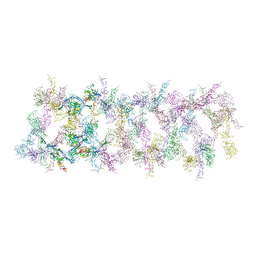 | | Netrin-1 filament assembly | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Netrin-1, ... | | Authors: | McDougall, M, Gupta, M, Stetefeld, J. | | Deposit date: | 2021-01-14 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (5.99 Å) | | Cite: | The dynamic nature of netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation.
Nat Commun, 14, 2023
|
|
9IU1
 
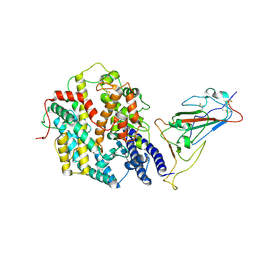 | | Structure of SARS-CoV-2 JN.1 spike RBD in complex with ACE2 (up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-07-20 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
1JF0
 
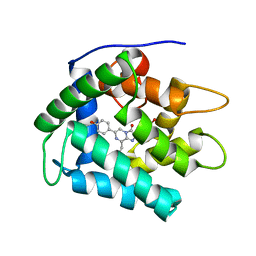 | | The Crystal Structure of Obelin from Obelia geniculata at 1.82 A Resolution | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, Obelin | | Authors: | Deng, L, Vysotski, E, Liu, Z.-J, Markova, S, Lee, J, Rose, J, Wang, B.-C. | | Deposit date: | 2001-06-19 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structure of Obelin from Obelia geniculata at 1.82 Angstrom Resolution
To be Published
|
|
7CHV
 
 | | Metallo-Beta-Lactamase VIM-2 in complex with 1-benzyl-1H-imidazole-2-carboxylic acid | | Descriptor: | 1-(phenylmethyl)imidazole-2-carboxylic acid, Beta-lactamase class B VIM-2, FORMIC ACID, ... | | Authors: | Yan, Y.-H, Li, G.-B. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | AncPhore: A versatile tool for anchor pharmacophore steered drug discovery with applications in discovery of new inhibitors targeting metallo-beta-lactamases and indoleamine/tryptophan 2,3-dioxygenases.
Acta Pharm Sin B, 11, 2021
|
|
4YRJ
 
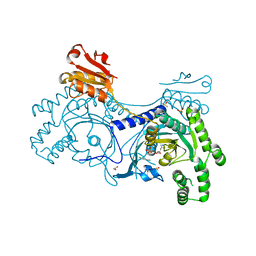 | | Crystal structure of T. cruzi Histidyl-tRNA synthetase in complex with 4-chlorobenzene-1,2-diamine (Chem 256) | | Descriptor: | 1,2-ETHANEDIOL, 4-chlorobenzene-1,2-diamine, DIMETHYL SULFOXIDE, ... | | Authors: | Koh, C.-Y, Hol, W.G.J. | | Deposit date: | 2015-03-15 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A binding hotspot in Trypanosoma cruzi histidyl-tRNA synthetase revealed by fragment-based crystallographic cocktail screens.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
5ZOG
 
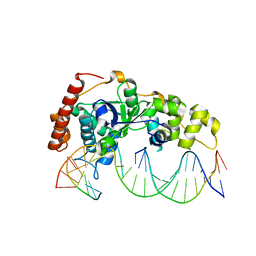 | | Crystal Structure of R192F hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
4OPM
 
 | |
3D4R
 
 | |
1JGM
 
 | | High Resolution Structure of the Cadmium-containing Phosphotriesterase from Pseudomonas diminuta | | Descriptor: | 1,2-ETHANEDIOL, 2-PHENYL-ETHANOL, CADMIUM ION, ... | | Authors: | Benning, M.M, Shim, H, Raushel, F.M, Holden, H.M. | | Deposit date: | 2001-06-26 | | Release date: | 2001-07-04 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High resolution X-ray structures of different metal-substituted forms of phosphotriesterase from Pseudomonas diminuta.
Biochemistry, 40, 2001
|
|
3KZ0
 
 | | MCL-1 complex with MCL-1-specific selected peptide | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Mcl-1 specific peptide MB7, SULFATE ION, ... | | Authors: | Dutta, S, Fire, E, Grant, R.A, Sauer, R.T, Keating, A.E. | | Deposit date: | 2009-12-07 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL.
J.Mol.Biol., 398, 2010
|
|
3KFS
 
 | |
4MCC
 
 | | HinTrmD in complex with N-[4-(AMINOMETHYL)BENZYL]-4-OXO-3,4-DIHYDROTHIENO[2,3-D]PYRIMIDINE-5-CARBOXAMIDE | | Descriptor: | N-[4-(aminomethyl)benzyl]-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Olivier, N.B, Hill, P. | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selective Inhibitors of Bacterial t-RNA-(N(1)G37) Methyltransferase (TrmD) That Demonstrate Novel Ordering of the Lid Domain.
J.Med.Chem., 56, 2013
|
|
6DBN
 
 | | Jak1 with compound 23 | | Descriptor: | Tyrosine-protein kinase JAK1, [(1S)-2,2-difluorocyclopropyl][(1R,5S)-3-{2-[(1-methyl-1H-pyrazol-4-yl)amino]pyrimidin-4-yl}-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Dual Inhibition of TYK2 and JAK1 for the Treatment of Autoimmune Diseases: Discovery of (( S)-2,2-Difluorocyclopropyl)((1 R,5 S)-3-(2-((1-methyl-1 H-pyrazol-4-yl)amino)pyrimidin-4-yl)-3,8-diazabicyclo[3.2.1]octan-8-yl)methanone (PF-06700841).
J. Med. Chem., 61, 2018
|
|
