2IS1
 
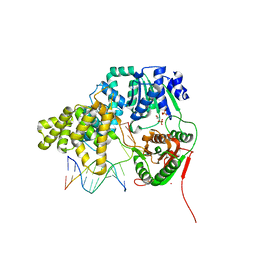 | | Crystal structure of UvrD-DNA-SO4 complex | | Descriptor: | 5'-D(*CP*GP*AP*GP*CP*AP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*TP*GP*CP*TP*CP*GP*TP*TP*TP*TP*TP*TP*T)-3', DNA helicase II, ... | | Authors: | Yang, W, Lee, J.Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke.
Cell(Cambridge,Mass.), 127, 2006
|
|
2FU3
 
 | |
2IS6
 
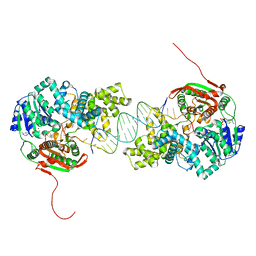 | | Crystal structure of UvrD-DNA-ADPMgF3 ternary complex | | Descriptor: | 5'-D(*CP*GP*AP*GP*CP*AP*CP*TP*GP*CP*AP*GP*TP*GP*CP*TP*CP*GP*TP*TP*GP*TP*TP*AP*T)-3', ADENOSINE-5'-DIPHOSPHATE, DNA helicase II, ... | | Authors: | Yang, W, Lee, J.Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke.
Cell(Cambridge,Mass.), 127, 2006
|
|
2IJM
 
 | |
2IRU
 
 | |
2IEJ
 
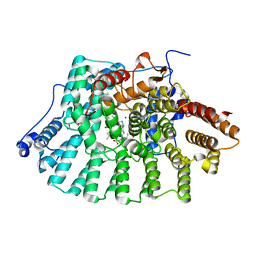 | | Human Protein Farnesyltransferase Complexed with Inhibitor Compound STN-48 And FPP Analog at 1.8A Resolution | | Descriptor: | ACETATE ION, METHYL N-{(3S)-1-[(1-METHYL-1H-IMIDAZOL-5-YL)METHYL]-6-PHENYL-1,2,3,4-TETRAHYDROQUINOLIN-3-YL}-N-[(1-METHYL-1H-IMIDAZOL-4-YL)SULFONYL]GLYCINATE, Protein farnesyltransferase subunit beta, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-01-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Resistance mutations at the lipid substrate binding site of Plasmodium falciparum protein farnesyltransferase.
Mol.Biochem.Parasitol., 152, 2007
|
|
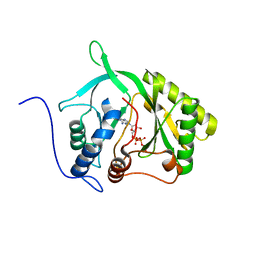
2IRY
 
 | |
2IS4
 
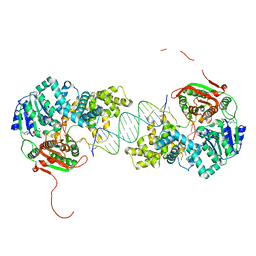 | | Crystal structure of UvrD-DNA-ADPNP ternary complex | | Descriptor: | 25-MER, DNA helicase II, MAGNESIUM ION, ... | | Authors: | Yang, W, Lee, J.Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke.
Cell(Cambridge,Mass.), 127, 2006
|
|
8J2E
 
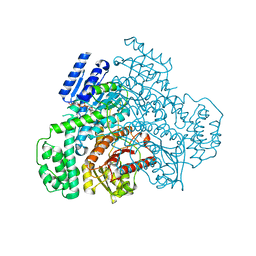 | |
8QK7
 
 | | E167K RF2 on E. coli 70S release complex with UAA | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Pundir, S, Larsson, D.S.D, Selmer, M, Sanyal, S. | | Deposit date: | 2023-09-14 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | The compensatory mechanism of a naturally evolved
E167K RF2 counteracting the loss of
RF1 in bacteria
To Be Published
|
|
8J2B
 
 | |
2K7Y
 
 | |
8QKU
 
 | | SWR1-nucleosome complex in configuration 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Jalal, A.S.B, Wigley, D.B. | | Deposit date: | 2023-09-18 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Nucleosome flipping drives kinetic proof reading and processivity of histone exchange by yeast SWR1 complex
To Be Published
|
|
2K9E
 
 | |
2EAV
 
 | |
8QN8
 
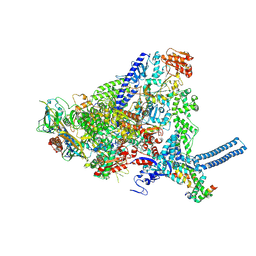 | | Mycobacterium smegmatis RNA polymerase in complex with HelD, SigA and RbpA in State II | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Koval, T, Krasny, L, Dohnalek, J, Kouba, T. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Molecular insights into RNA polymerase recycling by mycobacterial HelD
To Be Published
|
|
8QU6
 
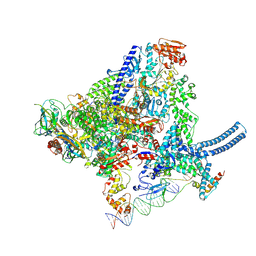 | | Mycobacterium smegnatis RNA polymerase transcription initiation complex with SigmaA, RbpA, HelD and an upstream-fork promoter fragment | | Descriptor: | DNA 22-MER, DNA 31-MER, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Koval, T, Krasny, L, Dohnalek, J, Kouba, T. | | Deposit date: | 2023-10-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Molecular insights into RNA polymerase recycling by mycobacterial HelD
To Be Published
|
|
8QKV
 
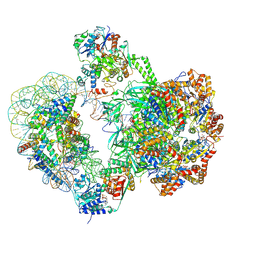 | | SWR1-nucleosome complex in configuration 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Jalal, A.S.B, Wigley, D.B. | | Deposit date: | 2023-09-18 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Nucleosome flipping drives kinetic proof reading and processivity of histone exchange by yeast SWR1 complex
To Be Published
|
|
2K6D
 
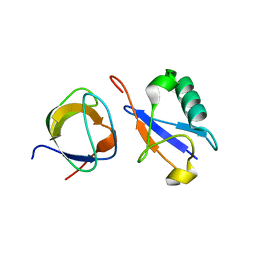 | | CIN85 Sh3-C domain in complex with ubiquitin | | Descriptor: | SH3 domain-containing kinase-binding protein 1, Ubiquitin | | Authors: | Forman-Kay, J, Bezsonova, I. | | Deposit date: | 2008-07-07 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Interactions between the three CIN85 SH3 domains and ubiquitin: implications for CIN85 ubiquitination.
Biochemistry, 47, 2008
|
|
8ISO
 
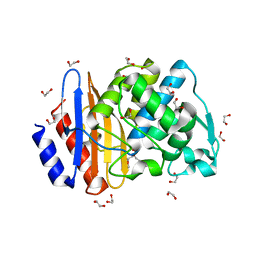 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
1FV1
 
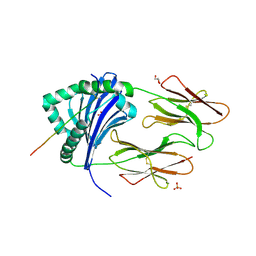 | | STRUCTURAL BASIS FOR THE BINDING OF AN IMMUNODOMINANT PEPTIDE FROM MYELIN BASIC PROTEIN IN DIFFERENT REGISTERS BY TWO HLA-DR2 ALLELES | | Descriptor: | GLYCEROL, MAJOR HISTOCOMPATIBILITY COMPLEX ALPHA CHAIN, MAJOR HISTOCOMPATIBILITY COMPLEX BETA CHAIN, ... | | Authors: | Li, H, Mariuzza, A.R, Li, Y, Martin, R. | | Deposit date: | 2000-09-18 | | Release date: | 2000-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of an immunodominant peptide from myelin basic protein in different registers by two HLA-DR2 proteins.
J.Mol.Biol., 304, 2000
|
|
8ISP
 
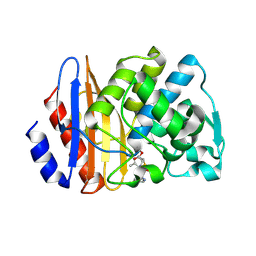 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cephalexin | | Descriptor: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISQ
 
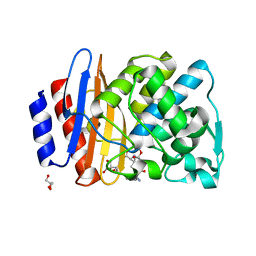 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
2K9G
 
 | |
8ISR
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cefaclor | | Descriptor: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-chloro-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
