1LFZ
 
 | | OXY HEMOGLOBIN (25% METHANOL) | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Biswal, B.K, Vijayan, M. | | Deposit date: | 2002-04-12 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of human oxy- and deoxyhaemoglobin at different levels of humidity: variability in the T state.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1HR2
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A MUTANT P4-P6 DOMAIN (DELC209) OF TETRAHYMENA THEMOPHILA GROUP I INTRON. | | Descriptor: | MAGNESIUM ION, P4-P6 DELC209 MUTANT RNA RIBOZYME DOMAIN | | Authors: | Juneau, K, Podell, E.R, Harrington, D.J, Cech, T.R. | | Deposit date: | 2000-12-20 | | Release date: | 2001-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA--solvent interactions.
Structure, 9, 2001
|
|
2GMJ
 
 | | Structure of Porcine Electron Transfer Flavoprotein-Ubiquinone Oxidoreductase | | Descriptor: | Electron transfer flavoprotein-ubiquinone oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Zhang, J, Frerman, F.E, Kim, J.-J.P. | | Deposit date: | 2006-04-06 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1JX4
 
 | | Crystal Structure of a Y-family DNA Polymerase in a Ternary Complex with DNA Substrates and an Incoming Nucleotide | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-DIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*A)-3', 5'-D(*T*TP*CP*AP*TP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Ling, H, Boudsocq, F, Woodgate, R, Yang, W. | | Deposit date: | 2001-09-05 | | Release date: | 2001-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication.
Cell(Cambridge,Mass.), 107, 2001
|
|
1JFI
 
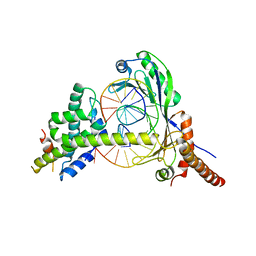 | | Crystal Structure of the NC2-TBP-DNA Ternary Complex | | Descriptor: | 5'-D(*G*GP*AP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*TP*CP*C)-3', TATA-BOX-BINDING PROTEIN (TBP), ... | | Authors: | Kamada, K, Shu, F, Chen, H, Malik, S, Stelzer, G, Roeder, R.G, Meisterernst, M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-06-20 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex.
Cell(Cambridge,Mass.), 106, 2001
|
|
1BLD
 
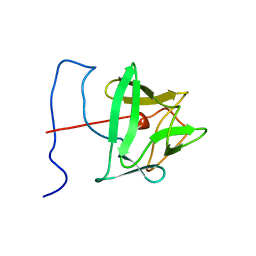 | | BASIC FIBROBLAST GROWTH FACTOR (FGF-2) MUTANT WITH CYS 78 REPLACED BY SER AND CYS 96 REPLACED BY SER, NMR | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Powers, R, Seddon, A.P, Bohlen, P, Moy, F.J. | | Deposit date: | 1996-05-20 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of basic fibroblast growth factor determined by multidimensional heteronuclear magnetic resonance spectroscopy.
Biochemistry, 35, 1996
|
|
1JXL
 
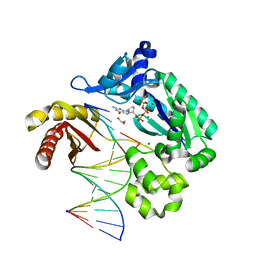 | | Crystal Structure of a Y-Family DNA Polymerase in a Ternary Complex with DNA Substrates and an Incoming Nucleotide | | Descriptor: | 1,2-ETHANEDIOL, 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*T)-3', ... | | Authors: | Ling, H, Boudsocq, F, Woodgate, R, Yang, W. | | Deposit date: | 2001-09-07 | | Release date: | 2001-10-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication.
Cell(Cambridge,Mass.), 107, 2001
|
|
1IIB
 
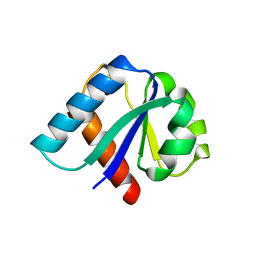 | | CRYSTAL STRUCTURE OF IIBCELLOBIOSE FROM ESCHERICHIA COLI | | Descriptor: | ENZYME IIB OF THE CELLOBIOSE-SPECIFIC PHOSPHOTRANSFERASE SYSTEM | | Authors: | Van Montfort, R.L.M, Pijning, T, Kalk, K.H, Reizer, J, Saier, M.H, Thunnissen, M.M.G.M, Robillard, G.T, Dijkstra, B.W. | | Deposit date: | 1996-12-23 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of an energy-coupling protein from bacteria, IIBcellobiose, reveals similarity to eukaryotic protein tyrosine phosphatases.
Structure, 5, 1997
|
|
1DLQ
 
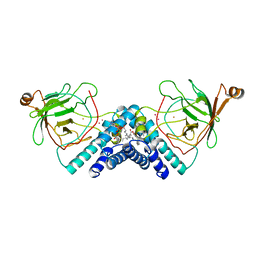 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 INHIBITED BY BOUND MERCURY | | Descriptor: | CATECHOL 1,2-DIOXYGENASE, FE (III) ION, MERCURY (II) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-11 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1CCV
 
 | |
1A61
 
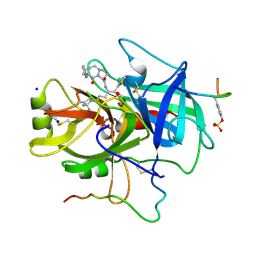 | | THROMBIN COMPLEXED WITH A BETA-MIMETIC THIAZOLE-CONTAINING INHIBITOR | | Descriptor: | (1S,7S)-7-amino-7-benzyl-N-{(1S)-4-carbamimidamido-1-[(S)-hydroxy(1,3-thiazol-2-yl)methyl]butyl}-8-oxohexahydro-1H-pyra zolo[1,2-a]pyridazine-1-carboxamide, ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), ... | | Authors: | St Charles, R, Matthews, J.H, Zhang, E, Tulinsky, A, Kahn, M. | | Deposit date: | 1998-03-05 | | Release date: | 1998-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bound structures of novel P3-P1' beta-strand mimetic inhibitors of thrombin.
J.Med.Chem., 42, 1999
|
|
2MG9
 
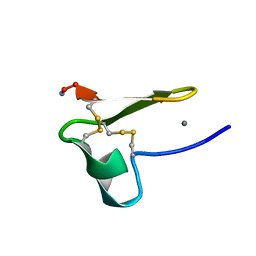 | | Truncated EGF-A | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor | | Authors: | Schroeder, C.I, Rosengren, K. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Design and Synthesis of Truncated EGF-A Peptides that Restore LDL-R Recycling in the Presence of PCSK9 In Vitro.
Chem.Biol., 21, 2014
|
|
1A46
 
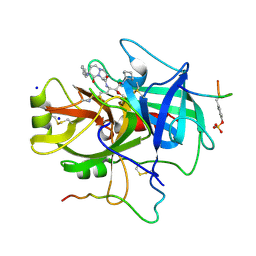 | | THROMBIN COMPLEXED WITH HIRUGEN AND A BETA-STRAND MIMETIC INHIBITOR | | Descriptor: | (1S,7S)-7-amino-N-[(2R,3S)-7-amino-1-(cyclohexylamino)-2-hydroxy-1-oxoheptan-3-yl]-7-benzyl-8-oxohexahydro-1H-pyrazolo[1,2-a]pyridazine-1-carboxamide, ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), ... | | Authors: | St Charles, R, Matthews, J.H, Zhang, E, Tulinsky, A, Kahn, M. | | Deposit date: | 1998-02-11 | | Release date: | 1998-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Bound structures of novel P3-P1' beta-strand mimetic inhibitors of thrombin.
J.Med.Chem., 42, 1999
|
|
2F2K
 
 | | Aldose reductase tertiary complex with NADPH and DEG | | Descriptor: | Aldose reductase, GAMMA-GLUTAMYL-S-(1,2-DICARBOXYETHYL)CYSTEINYLGLYCINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Singh, R, White, M.A, Ramana, K.V, Petrash, J.M, Watowich, S.J, Bhatnagar, A, Srivastava, S.K. | | Deposit date: | 2005-11-17 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of a glutathione conjugate bound to the active site of aldose reductase.
Proteins, 64, 2006
|
|
1DLT
 
 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 WITH BOUND CATECHOL | | Descriptor: | CATECHOL, CATECHOL 1,2-DIOXYGENASE, FE (III) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-12 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
2JTT
 
 | |
2G1Q
 
 | | crystal structure of KSP in complex with inhibitor 9h | | Descriptor: | (5S)-5-(3-AMINOPROPYL)-3-(2,5-DIFLUOROPHENYL)-N-ETHYL-5-PHENYL-4,5-DIHYDRO-1H-PYRAZOLE-1-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2006-02-14 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 4: Structure-based design of 5-alkylamino-3,5-diaryl-4,5-dihydropyrazoles as potent, water-soluble inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1JOI
 
 | | STRUCTURE OF PSEUDOMONAS FLUORESCENS HOLO AZURIN | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Lee, X, Ton-that, H, Zhu, D.-W, Biesterfedlt, J, Lanthier, P.H, Yachuchi, M, Dahms, T, Szabo, A.G. | | Deposit date: | 1997-06-09 | | Release date: | 1997-12-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallization and preliminary crystallographic studies of the crystals of the azurin Pseudomonas fluorescens.
Arch.Biochem.Biophys., 308, 1994
|
|
1DTT
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 (PETT130A94) | | Descriptor: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-01-13 | | Release date: | 2000-04-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1DTQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-1 (PETT131A94) | | Descriptor: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-NITRILOMETHYL-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1EAS
 
 | | NONPEPTIDIC INHIBITORS OF HUMAN LEUKOCYTE ELASTASE. 3. DESIGN, SYNTHESIS, X-RAY CRYSTALLOGRAPHIC ANALYSIS, AND STRUCTURE-ACTIVITY RELATIONSHIPS FOR A SERIES OF ORALLY ACTIVE 3-AMINO-6-PHENYLPYRIDIN-2-ONE TRIFLUOROMETHYL KETONES | | Descriptor: | 3-[[(METHYLAMINO)SULFONYL]AMINO]-2-OXO-6-PHENYL-N-[3,3,3-TRIFLUORO-1-(1-METHYLETHYL)-2-OXOPHENYL]-1(2H)-PYRIDINE ACETAMIDE, PORCINE PANCREATIC ELASTASE, SODIUM ION, ... | | Authors: | Ceccarelli, C. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nonpeptidic inhibitors of human leukocyte elastase. 3. Design, synthesis, X-ray crystallographic analysis, and structure-activity relationships for a series of orally active 3-amino-6-phenylpyridin-2-one trifluoromethyl ketones.
J.Med.Chem., 37, 1994
|
|
1JBZ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A DUAL-WAVELENGTH EMISSION GREEN FLUORESCENT PROTEIN VARIANT AT HIGH PH | | Descriptor: | 1,2-ETHANEDIOL, GREEN FLUORESCENT PROTEIN, MAGNESIUM ION | | Authors: | Hanson, G.T, McAnaney, T.B, Park, E.S, Rendell, M.E.P, Yarbrough, D.K, Chu, S, Xi, L, Boxer, S.G, Montrose, M.H, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Green Fluorescent Protein Variants as Ratiometric Dual Emission pH Sensors. 1. Structural Characterization and Preliminary Application.
Biochemistry, 41, 2002
|
|
1TLF
 
 | |
1JBY
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A DUAL-WAVELENGTH EMISSION GREEN FLUORESCENT PROTEIN VARIANT AT LOW PH | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Hanson, G.T, McAnaney, T.B, Park, E.S, Rendell, M.E.P, Yarbrough, D.K, Chu, S, Xi, L, Boxer, S.G, Montrose, M.H, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Green Fluorescent Protein Variants as Ratiometric Dual Emission pH Sensors. 1. Structural Characterization and Preliminary Application.
Biochemistry, 41, 2002
|
|
1DMH
 
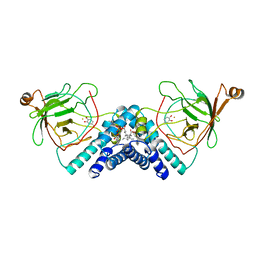 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 WITH BOUND 4-METHYLCATECHOL | | Descriptor: | 4-METHYLCATECHOL, CATECHOL 1,2-DIOXYGENASE, FE (III) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
