2PRZ
 
 | |
4C69
 
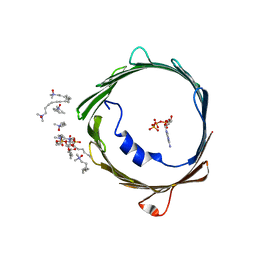 | | ATP binding to murine voltage-dependent anion channel 1 (mVDAC1). | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Paz, A, Colletier, J.P, Abramson, J. | | Deposit date: | 2013-09-17 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.277 Å) | | Cite: | Structure-Guided Simulations Illuminate the Mechanism of ATP Transport Through Vdac1.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1K0Y
 
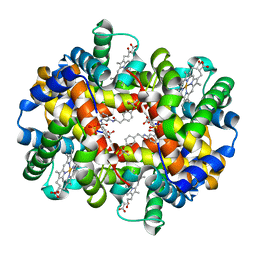 | | X-ray Crystallographic Analyses of Symmetrical Allosteric Effectors of Hemoglobin. Compounds Designed to Link Primary and Secondary Binding Sites | | Descriptor: | 2-{4-[(3{2-[4-(1-CARBOXY-1-METHYL-ETHOXY)-PHENYL]-ACETYLAMINO}-PHENYLCARBAMOYL)-METHYL]-PHENOXY}-2-METHYL-PROPIONIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Safo, M.K, Boyiri, T, Burnett, J.C, Danso-Danquah, R, Moure, C.M, Joshi, G.S, Abraham, D.J. | | Deposit date: | 2001-09-21 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | X-ray crystallographic analyses of symmetrical allosteric effectors of hemoglobin: compounds designed to link primary and secondary binding sites.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5GMZ
 
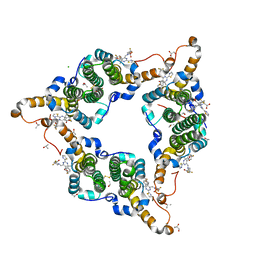 | | Hepatitis B virus core protein Y132A mutant in complex with 4-methyl heteroaryldihydropyrimidine | | Descriptor: | (2S)-4,4-difluoro-1-[[(4S)-4-(4-fluorophenyl)-5-methoxycarbonyl-4-methyl-2-(1,3-thiazol-2-yl)-1H-pyrimidin-6-yl]methyl]pyrrolidine-2-carboxylic acid, CHLORIDE ION, Core protein, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Synthesis of Orally Bioavailable 4-Methyl Heteroaryldihydropyrimidine Based Hepatitis B Virus (HBV) Capsid Inhibitors
J.Med.Chem., 59, 2016
|
|
7PC6
 
 | | DNA-binding domain of a p53 homolog from the hydrothermal vent annelid Alvinella pompejana | | Descriptor: | 1,2-ETHANEDIOL, DNA-binding domain, ZINC ION | | Authors: | Balourdas, D.-I, Knapp, S, Soussi, T, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Evolutionary history of the p53 family DNA-binding domain: insights from an Alvinella pompejana homolog.
Cell Death Dis, 13, 2022
|
|
1UNS
 
 | | IDENTIFICATION OF A SECONDARY ZINC-BINDING SITE IN STAPHYLOCOCCAL ENTEROTOXIN C2: IMPLICATIONS FOR SUPERANTIGEN RECOGNITION | | Descriptor: | ENTEROTOXIN TYPE C-2, ZINC ION | | Authors: | Papageorgiou, A.C, Baker, M.D, McLeod, J.D, Goda, S, Sansom, D.M, Tranter, H.S, Acharya, K.R. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a Secondary Zinc-Binding Site in Staphylococcal Enterotoxin C2: Implications for Superantigen Recognition
J.Biol.Chem., 279, 2004
|
|
8SF8
 
 | | Structure of bovine PKA bound to (R)-N-(4-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl)-2-amino-4-methylpentanamide | | Descriptor: | N-[4-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl]-D-leucinamide, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Coker, J.A, Arya, T, Goins, C.M, Maw, J.J, Macdonald, J.D, Stauffer, S.R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Characterization of Selective, First-in-Class Inhibitors of Citron Kinase.
J.Med.Chem., 67, 2024
|
|
3IG8
 
 | |
3IG5
 
 | |
4B2T
 
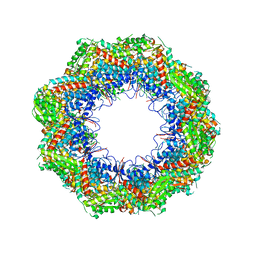 | | The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT ALPHA, T-COMPLEX PROTEIN 1 SUBUNIT BETA, T-COMPLEX PROTEIN 1 SUBUNIT DELTA, ... | | Authors: | Kalisman, N, Schroeder, G.F, Levitt, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | The Crystal Structures of the Eukaryotic Chaperonin Cct Reveal its Functional Partitioning
Structure, 21, 2013
|
|
6MBE
 
 | | Human Mcl-1 in complex with the designed peptide dM7 | | Descriptor: | CHLORIDE ION, Induced myeloid leukemia cell differentiation protein Mcl-1, dM7 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6MBC
 
 | | Human Bfl-1 in complex with the designed peptide dF4 | | Descriptor: | Bcl-2-related protein A1, dF4 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6MBD
 
 | | Human Mcl-1 in complex with the designed peptide dM1 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION, dM1 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
8SHI
 
 | | Valpha3S1 Vbeta13S1 HLA C 0602 VRSRRCLRL | | Descriptor: | Beta-2-microglobulin, MHC class I antigen (Fragment), T cell receptor alpha, ... | | Authors: | Littler, D.R, Anand, S, Vivian, J.P, Rossjohn, J. | | Deposit date: | 2023-04-14 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.90001726 Å) | | Cite: | Complimentary electrostatics dominate T-cell receptor binding to a psoriasis-associated peptide antigen presented by human leukocyte antigen C∗06:02.
J.Biol.Chem., 299, 2023
|
|
8JZC
 
 | | Crystal structure of Geobacillus stearothermophilus NarJ | | Descriptor: | Nitrate reductase molybdenum cofactor assembly chaperone | | Authors: | Song, W.S, Kim, J.H, Namgung, B, Cho, H.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-07-05 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Complementary hydrophobic interaction of the redox enzyme maturation protein NarJ with the signal peptide of the respiratory nitrate reductase NarG.
Int.J.Biol.Macromol., 262, 2024
|
|
8JZD
 
 | | Crystal structure of Escherichia coli NarJ in complex with the signal peptide of E. coli NarG | | Descriptor: | Nitrate reductase molybdenum cofactor assembly chaperone NarJ, Respiratory nitrate reductase 1 alpha chain | | Authors: | Song, W.S, Kim, J.H, Namgung, B, Cho, H.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-07-05 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Complementary hydrophobic interaction of the redox enzyme maturation protein NarJ with the signal peptide of the respiratory nitrate reductase NarG.
Int.J.Biol.Macromol., 262, 2024
|
|
7OCI
 
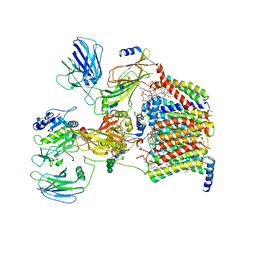 | | Cryo-EM structure of yeast Ost6p containing oligosaccharyltransferase complex | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wild, R, Neuhaus, J.D, Eyring, J, Irobalieva, R.N, Kowal, J, Lin, C.W, Locher, K.P, Aebi, M. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Functional analysis of Ost3p and Ost6p containing yeast oligosaccharyltransferases.
Glycobiology, 31, 2021
|
|
4JBZ
 
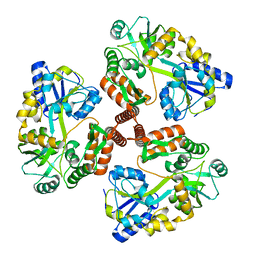 | | Structure of Mcm10 coiled-coil region | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN FUSED WITH XENOPUS LAEVIS MCM10 COILED-COIL REGION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Du, W, Adhikary, S, Eichman, B.F. | | Deposit date: | 2013-02-20 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mcm10 self-association is mediated by an N-terminal coiled-coil domain.
Plos One, 8, 2013
|
|
6O3R
 
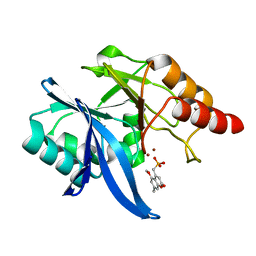 | | Crystal Structure of NDM-1 D199N with Compound 7 | | Descriptor: | Carbapenem Hydrolyzing Class B Metallo beta lactamase NDM-1, ZINC ION, [(5-methoxy-7-methyl-2-oxo-2H-1-benzopyran-4-yl)methyl]phosphonic acid | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Heteroaryl Phosphonates as Noncovalent Inhibitors of Both Serine- and Metallocarbapenemases.
J.Med.Chem., 62, 2019
|
|
6NY7
 
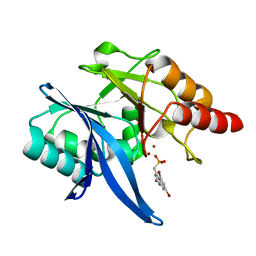 | | Crystal Structure of NDM-1 D199N with Compound 16 | | Descriptor: | Carbapenem Hydrolyzing Class B Metallo beta lactamase NDM-1, ZINC ION, [(5,7-dibromo-2-oxo-1,2-dihydroquinolin-4-yl)methyl]phosphonic acid | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2019-02-11 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Heteroaryl Phosphonates as Noncovalent Inhibitors of Both Serine- and Metallocarbapenemases.
J.Med.Chem., 62, 2019
|
|
5TJ5
 
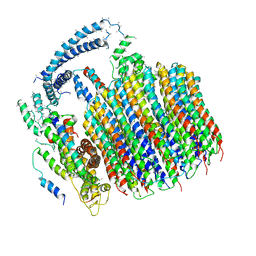 | | Atomic model for the membrane-embedded motor of a eukaryotic V-ATPase | | Descriptor: | V-type proton ATPase subunit a, V-type proton ATPase subunit c, V-type proton ATPase subunit c', ... | | Authors: | Mazhab-Jafari, M.T, Rohou, A, Schmidt, C, Bueler, S.A, Benlekbir, S, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2016-10-03 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase.
Nature, 539, 2016
|
|
2ENB
 
 | |
1HBZ
 
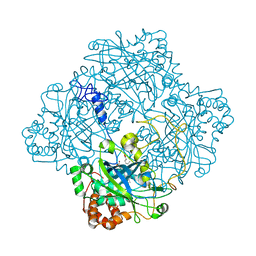 | | Catalase from Micrococcus lysodeikticu | | Descriptor: | CATALASE, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Murshudov, G.N, Grebenko, A.I, Barynin, V.V, Braningen, J, Wilson, K.S, Dauter, Z, Melik-Adamyan, W.R. | | Deposit date: | 2001-04-24 | | Release date: | 2001-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-Dimensional Structure of Catalase from Micrococcus Lysodeikticus at 1.5A Resolution
FEBS Lett., 312, 1992
|
|
7R4L
 
 | | Crystal structure of human mitochondrial NAD kinase | | Descriptor: | FE (III) ION, NAD kinase 2, mitochondrial, ... | | Authors: | Labesse, G, Mary, C, Gelin, M, Lionne, C. | | Deposit date: | 2022-02-08 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human NADK2 reveals a dimeric organization and active site occlusion by lysine acetylation.
Mol.Cell, 82, 2022
|
|
8RBL
 
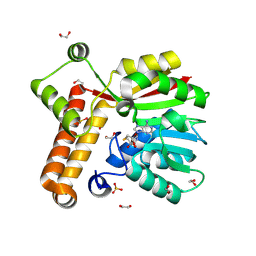 | | Crystal structure of Mycobacterium tuberculosis MmaA1 with S-adenosyl homocysteine | | Descriptor: | 1,2-ETHANEDIOL, Mycolic acid methyltransferase MmaA1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Kobakhidze, G, Wachelder, L, Chaudhary, B, Mazumdar, P.A, Madhurantakam, C, Dong, G. | | Deposit date: | 2023-12-04 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of the mycolic acid methyl transferase 1 (MmaA1) from Mycobacterium tuberculosis in the apo-form and in complex with different cofactors reveal unique features for substrate binding.
J.Biomol.Struct.Dyn., 2025
|
|
