2JR1
 
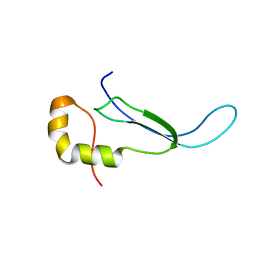 | | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from the phytopathogen Xylella fastidiosa. | | Descriptor: | Virulence regulator | | Authors: | Rosselli, L.K, Sforca, M.L, Souza, A.P, Zeri, A.C. | | Deposit date: | 2007-06-18 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from the phytopathogen Xylella fastidiosa.
To be Published
|
|
5A39
 
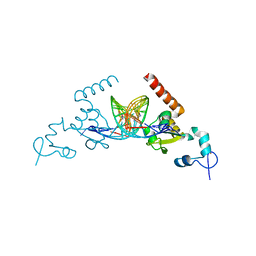 | | Structure of Rad14 in complex with cisplatin containing DNA | | Descriptor: | 5'-D(*DGP*AP*TP*GP*AP*CP*CP*GP*TP*AP*GP*AP*GP)-3', 5'-D(*DTP*GP*AP*TP*GP*AP*CP*CP*GP*TP*AP*GP*AP)-3', Cisplatin, ... | | Authors: | Koch, S.C, Kuper, J, Gasteiger, K.L, Wichlein, N, Strasser, R, Eisen, D, Geiger, S, Schneider, S, Kisker, C, Carell, T. | | Deposit date: | 2015-05-28 | | Release date: | 2015-06-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into the Recognition of Cisplatin and Aaf-Dg Lesion by Rad14 (Xpa).
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6I89
 
 | |
6GRD
 
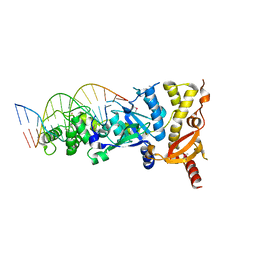 | | eukaryotic junction-resolving enzyme GEN-1 binding with Cesium | | Descriptor: | CESIUM ION, DNA (5'-D(*TP*AP*CP*CP*CP*AP*CP*CP*AP*CP*CP*GP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), ... | | Authors: | Lilley, D.M.J, Liu, Y, Freeman, D.J. | | Deposit date: | 2018-06-11 | | Release date: | 2018-09-26 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.
Nucleic Acids Res., 46, 2018
|
|
1ZTT
 
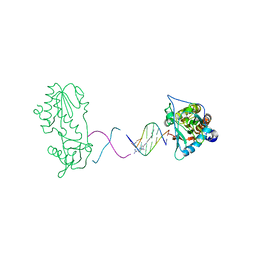 | | Netropsin bound to d(CTTAATTCGAATTAAG) in complex with MMLV RT catalytic fragment | | Descriptor: | 5'-D(*CP*TP*TP*AP*AP*TP*TP*C)-3', 5'-D(P*GP*AP*AP*TP*TP*AP*AP*G)-3', NETROPSIN, ... | | Authors: | Goodwin, K.D, Long, E.C, Georgiadis, M.M. | | Deposit date: | 2005-05-27 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A host-guest approach for determining drug-DNA interactions: an example using netropsin.
Nucleic Acids Res., 33, 2005
|
|
6GVW
 
 | | Crystal structure of the BRCA1-A complex | | Descriptor: | BRCA1-A complex subunit Abraxas 1, BRCA1-A complex subunit RAP80, BRISC and BRCA1-A complex member 1, ... | | Authors: | Bunker, R.D, Rabl, J, Thoma, N.H. | | Deposit date: | 2018-06-21 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structural Basis of BRCC36 Function in DNA Repair and Immune Regulation.
Mol.Cell, 75, 2019
|
|
5ZD4
 
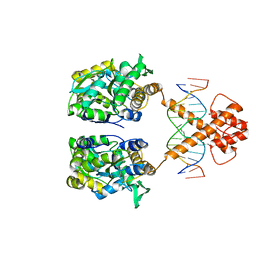 | | Crystal structure of MBP-fused BIL1/BZR1 in complex with double-stranded DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*GP*TP*GP*TP*GP*AP*AP*A)-3'), Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Miyakawa, T, Xu, Y, Nakamura, A, Hirabayashi, K, Tanokura, M. | | Deposit date: | 2018-02-22 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis for brassinosteroid response by BIL1/BZR1.
Nat Plants, 4, 2018
|
|
6H3C
 
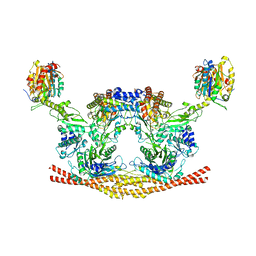 | | Cryo-EM structure of the BRISC complex bound to SHMT2 | | Descriptor: | BRISC and BRCA1-A complex member 1, BRISC and BRCA1-A complex member 2, BRISC complex subunit Abraxas 2, ... | | Authors: | Bunker, R.D, Rabl, J, Thoma, N.H. | | Deposit date: | 2018-07-18 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of BRCC36 Function in DNA Repair and Immune Regulation.
Mol.Cell, 75, 2019
|
|
2ABK
 
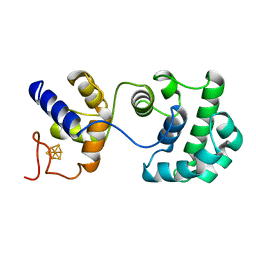 | |
2B1B
 
 | |
2LVH
 
 | | Solution structure of the zinc finger AFV1p06 protein from the hyperthermophilic archaeal virus AFV1 | | Descriptor: | Putative zinc finger protein ORF59a, ZINC ION | | Authors: | Guilliere, F, Sezonov, G, Prangishvili, D, Delepierre, M, Guijarro, J. | | Deposit date: | 2012-07-05 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an archaeal DNA binding protein with an eukaryotic zinc finger fold.
Plos One, 8, 2013
|
|
2MKG
 
 | |
4HPZ
 
 | |
1WG3
 
 | | Structural analysis of yeast nucleosome-assembly factor CIA1p | | Descriptor: | Anti-silencing protein 1 | | Authors: | Padmanabhan, B, Kataoka, K, Umehara, T, Adachi, N, Yokoyama, S, Horikoshi, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Similarity between Histone Chaperone Cia1p/Asf1p and DNA-Binding Protein NF-{kappa}B
J.Biochem.(Tokyo), 138, 2005
|
|
2MKF
 
 | |
4QLI
 
 | |
7CEI
 
 | | THE ENDONUCLEASE DOMAIN OF COLICIN E7 IN COMPLEX WITH ITS INHIBITOR IM7 PROTEIN | | Descriptor: | PROTEIN (COLICIN E7 IMMUNITY PROTEIN), ZINC ION | | Authors: | Ko, T.P, Liao, C.C, Ku, W.Y, Chak, K.F, Yuan, H.S. | | Deposit date: | 1998-09-17 | | Release date: | 1999-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein.
Structure Fold.Des., 7, 1999
|
|
3C6H
 
 | | Crystal Structure of the RB49 gp17 nuclease domain | | Descriptor: | MAGNESIUM ION, Terminase large subunit | | Authors: | Sun, S, Rossmann, M.G. | | Deposit date: | 2008-02-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces.
Cell(Cambridge,Mass.), 135, 2008
|
|
2B6C
 
 | | Predicted DNA alkylation repair enzyme from Enterococcus faecalis. | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Osipiuk, J, Hatzos, C, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-30 | | Release date: | 2005-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Xray structure of predicted DNA alkylation repair enzyme from Enterococcus faecalis.
To be Published
|
|
3OIP
 
 | | Crystal structure of Yeast telomere protein Cdc13 OB1 | | Descriptor: | Cell division control protein 13 | | Authors: | Sun, J, Yang, Y, Wan, K, Mao, N, Yu, T.Y, Lin, Y.C, DeZwaan, D.C, Freeman, B.C, Lin, J.J, Lue, N.F, Lei, M. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha.
Cell Res., 21, 2011
|
|
2F4M
 
 | | The Mouse PNGase-HR23 Complex Reveals a Complete Remodulation of the Protein-Protein Interface Compared to its Yeast Orthologs | | Descriptor: | CHLORIDE ION, UV excision repair protein RAD23 homolog B, ZINC ION, ... | | Authors: | Zhao, G, Zhou, X, Wang, L, Kisker, C, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2005-11-23 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the mouse peptide N-glycanase-HR23 complex suggests co-evolution of the endoplasmic reticulum-associated degradation and DNA repair pathways.
J.Biol.Chem., 281, 2006
|
|
2XYZ
 
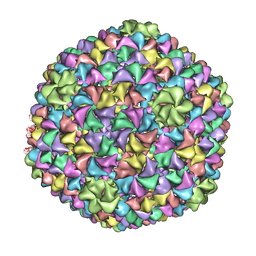 | | De Novo model of Bacteriophage P22 virion coat protein | | Descriptor: | COAT PROTEIN | | Authors: | Chen, D.-H, Baker, M.L, Hryc, C.F, DiMaio, F, Jakana, J, Wu, W, Dougherty, M, Haase-Pettingell, C, Schmid, M.F, Jiang, W, Baker, D, King, J.A, Chiu, W. | | Deposit date: | 2010-11-19 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis for Scaffolding-Mediated Assembly and Maturation of a DsDNA Virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3C6A
 
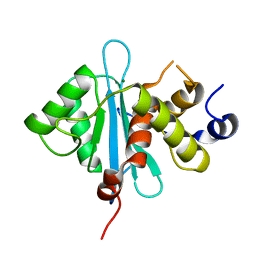 | | Crystal Structure of the RB49 gp17 nuclease domain | | Descriptor: | MAGNESIUM ION, Terminase large subunit | | Authors: | Sun, S, Rossmann, M.G. | | Deposit date: | 2008-02-04 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces.
Cell(Cambridge,Mass.), 135, 2008
|
|
2JXJ
 
 | |
2F4O
 
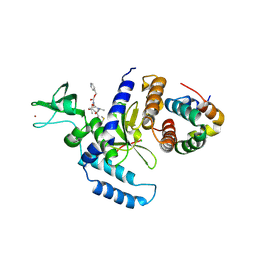 | | The Mouse PNGase-HR23 Complex Reveals a Complete Remodulation of the Protein-Protein Interface Compared to its Yeast Orthologs | | Descriptor: | CHLORIDE ION, PHQ-VAL-ALA-ASP-CF0, XP-C repair complementing complex 58 kDa protein, ... | | Authors: | Zhao, G, Zhou, X, Wang, L, Kisker, C, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2005-11-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of the mouse peptide N-glycanase-HR23 complex suggests co-evolution of the endoplasmic reticulum-associated degradation and DNA repair pathways.
J.Biol.Chem., 281, 2006
|
|
