6GVP
 
 | |
7XK9
 
 | | Structure of human beta2 adrenergic receptor bound to constrained isoproterenol | | Descriptor: | (5R,6R)-6-(propan-2-ylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Camelid Antibody Fragment, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Xu, X, Shonberg, J, Kaindl, J, Clark, M, Stobel, A, Maul, L, Mayer, D, Hubner, H, Venkatakrishnan, A, Dror, R, Kobilka, B.K, Sunahara, R, Liu, X, Gmeiner, P. | | Deposit date: | 2022-04-19 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Constrained catecholamines gain beta 2 AR selectivity through allosteric effects on pocket dynamics.
Nat Commun, 14, 2023
|
|
7KGP
 
 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N316-324 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
6GYA
 
 | | Amylase in complex with branched ligand | | Descriptor: | A-amylase, CALCIUM ION, SODIUM ION, ... | | Authors: | Agirre, J, Moroz, O, Meier, S, Brask, J, Munch, A, Hoff, T, Andersen, C, Wilson, K.S, Davies, G.J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The structure of the AliC GH13 alpha-amylase from Alicyclobacillus sp. reveals the accommodation of starch branching points in the alpha-amylase family.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7KUR
 
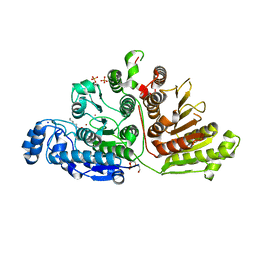 | |
7KSY
 
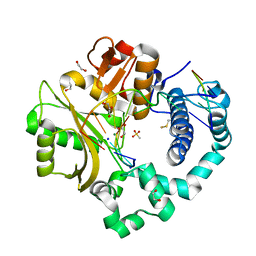 | | DNA Polymerase Mu, dGTP:Ct Product State Ternary Complex, 10 mM Mg2+ (960min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.577 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KSW
 
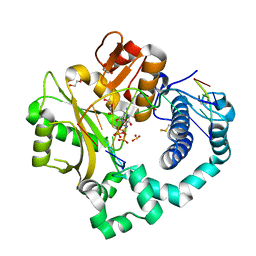 | | DNA Polymerase Mu, dGTP:Ct Reaction State Ternary Complex, 10 mM Mg2+ (10min) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KNK
 
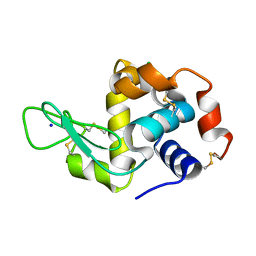 | |
7L6J
 
 | | Crystal Structure of the Putative Hydrolase from Stenotrophomonas maltophilia | | Descriptor: | CHLORIDE ION, FORMIC ACID, Putative hydrolase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the Putative Hydrolase from Stenotrophomonas maltophilia
To Be Published
|
|
7KSV
 
 | | DNA Polymerase Mu, dGTP:Ct Product State Ternary Complex, 10 mM Mn2+ (960min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KST
 
 | | DNA Polymerase Mu, dGTP:Ct Reaction State Ternary Complex, 10 mM Mn2+ (2min) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
6E7T
 
 | |
7XJV
 
 | | Crystal Structure of Alpha-1,3-mannosyltransferase MNT2 from Saccharomyces cerevisiae, Mn/GDP-mannose form | | Descriptor: | Alpha-1,3-mannosyltransferase MNT2, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Hira, D, Kadooka, C, Oka, T. | | Deposit date: | 2022-04-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Alpha-1,3-mannosyltransferase MNT2 from Saccharomyces cerevisiae, Mn/GDP-mannose form
To Be Published
|
|
7KT9
 
 | | DNA Polymerase Mu, 8-oxodGTP:At Product State Ternary Complex, 50 mM Mg2+ (960min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KXC
 
 | | The aminoacrylate form of the wild-type Salmonella typhimurium Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, sodium ion at the metal coordination site and benzimidazole (BZI) at the enzyme beta-site at 1.30 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-12-03 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The aminoacrylate form of the wild-type Salmonella typhimurium Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, sodium ion at the metal coordination site and benzimidazole (BZI) at the enzyme beta-site at 1.30 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring.
To be Published
|
|
7KT6
 
 | | DNA Polymerase Mu, 8-oxodGTP:At Product State Ternary Complex, 10 mM Mn2+ (960min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KTA
 
 | | DNA Polymerase Mu, 8-oxodGTP:Ct Pre-Catalytic Ground State Ternary Complex, 20 mM Ca2+ (120min) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KT5
 
 | | DNA Polymerase Mu, 8-oxodGTP:At Product State Ternary Complex, 10 mM Mn2+ (120min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KTF
 
 | | DNA Polymerase Mu, 8-oxodGTP:Ct Product State Ternary Complex, 50 mM Mg2+ (180min) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA (5'-D(*CP*GP*GP*CP*CP*TP*AP*CP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KT8
 
 | | DNA Polymerase Mu, 8-oxodGTP:At Product State Ternary Complex, 50 mM Mg2+ (180min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7XLD
 
 | | Crystal structure of IsdH linker-NEAT3 bound to a nanobody (VHH) | | Descriptor: | Iron-regulated surface determinant protein H, MAGNESIUM ION, Nanobody VHH6, ... | | Authors: | Caaveiro, J.M.M, Valenciano-Bellido, S, Tsumoto, K. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting hemoglobin receptors IsdH and IsdB of Staphylococcus aureus with a single VHH antibody inhibits bacterial growth.
J.Biol.Chem., 299, 2023
|
|
7XJB
 
 | | Rat-COMT, opicapone,SAM and Mg bond | | Descriptor: | CHLORIDE ION, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Takebe, K, Iijima, H, Suzuki, M, Kuwada-Kusunose, T. | | Deposit date: | 2022-04-15 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Computational Analyses of the Unique Interactions of Opicapone in the Binding Pocket of Catechol O -Methyltransferase: A Crystallographic Study and Fragment Molecular Orbital Analyses.
J.Chem.Inf.Model., 63, 2023
|
|
7KT3
 
 | | DNA Polymerase Mu, 8-oxodGTP:At Pre-Catalytic Ground State Ternary Complex, 20 mM Ca2+ (120min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KT4
 
 | | DNA Polymerase Mu, 8-oxodGTP:At Reaction State Ternary Complex, 10 mM Mn2+ (30min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KTN
 
 | | DNA Polymerase Mu, 8-oxodGTP:At Product State Ternary Complex, 10 mM Mg2+ (2160min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
