8VL9
 
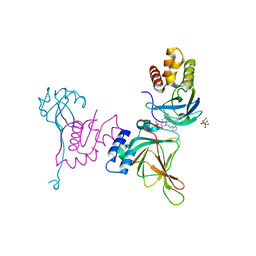 | | Crystal structure of EloBC-VHL-CDO1 complex bound to compound 8 molecular glue | | Descriptor: | CITRIC ACID, Cysteine dioxygenase type 1, Elongin-B, ... | | Authors: | Shu, W, Ma, X, Tutter, A, Buckley, D, Golosov, A, Michaud, G. | | Deposit date: | 2024-01-11 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A small molecule VHL molecular glue degrader for cysteine dioxygenase 1
To Be Published
|
|
8VL7
 
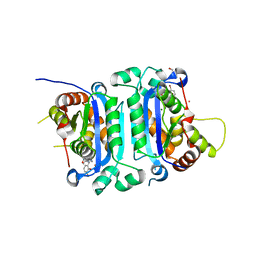 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | (2P)-2-[3-bromo-2-(2-hydroxyethoxy)phenyl]-5-hydroxy-1-methyl-N-(1,2-oxazol-4-yl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L. | | Deposit date: | 2024-01-11 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
8VKT
 
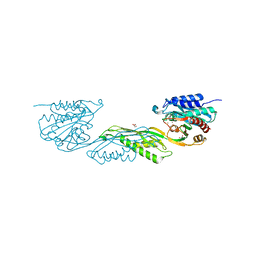 | | Crystallographic structure of dimetalated DapE from Enterococcus faecium | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Probable succinyl-diaminopimelate desuccinylase, ... | | Authors: | Gonzalez-Segura, L, Diaz-Vilchis, A, Terrazas-Lopez, M, Diaz-Sanchez, A.G. | | Deposit date: | 2024-01-09 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | The three-dimensional structure of DapE from Enterococcus faecium reveals new insights into DapE/ArgE subfamily ligand specificity.
Int.J.Biol.Macromol., 270, 2024
|
|
8VKF
 
 | |
8VKD
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrocatechol | | Descriptor: | 4-NITROCATECHOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8VKC
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrophenol | | Descriptor: | Dehaloperoxidase A, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8VKB
 
 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 10b | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, ... | | Authors: | Fenwick, M.K, Staker, B.L, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Exploring Subsite Selectivity within Plasmodium vivax N -Myristoyltransferase Using Pyrazole-Derived Inhibitors.
J.Med.Chem., 67, 2024
|
|
8VKA
 
 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 9c | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fenwick, M.K, Staker, B.L, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Exploring Subsite Selectivity within Plasmodium vivax N -Myristoyltransferase Using Pyrazole-Derived Inhibitors.
J.Med.Chem., 67, 2024
|
|
8VK2
 
 | | X-ray crystal structure of human IgE 4C8 Fab | | Descriptor: | IgE 4C8 heavy chain, IgE 4C8 light chain | | Authors: | Khatri, K, Ball, A, Smith, S.A, Champan, M.D, Pomes, A, Chruszcz, M. | | Deposit date: | 2024-01-08 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Structural analysis of human IgE monoclonal antibody epitopes on dust mite allergen Der p 2.
J.Allergy Clin.Immunol., 154, 2024
|
|
8VK1
 
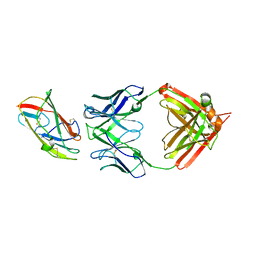 | | X-ray crystal structure of human IgE 4C8 Fab complex with Der p 2.0103 | | Descriptor: | Der p 2 variant 3, IgE 4C8 heavy chain, IgE 4C8 light chain | | Authors: | Khatri, K, Ball, A, Smith, S.A, Champan, M.D, Pomes, A, Chruszcz, M. | | Deposit date: | 2024-01-08 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural analysis of human IgE monoclonal antibody epitopes on dust mite allergen Der p 2.
J.Allergy Clin.Immunol., 154, 2024
|
|
8VJN
 
 | |
8VJ3
 
 | |
8VIS
 
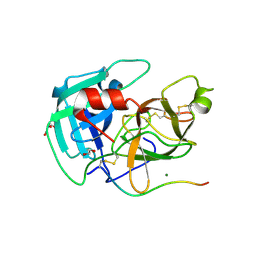 | | Human TMPRSS11D complexed with a disulfide-linked autoinhibitory DDDDK peptide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fraser, B.J, Dong, A, Ilyassov, O, Kenney, T, Li, Y.Y, Seitova, A, Li, Y, Hejazi, Z, Edwards, A, Benard, F, Arrowsmith, C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Human TMPRSS11D complexed with a disulfide-linked autoinhibitory DDDDK peptide
To Be Published
|
|
8VHU
 
 | |
8VHR
 
 | |
8VHQ
 
 | |
8VHP
 
 | |
8VHO
 
 | |
8VHN
 
 | |
8VHL
 
 | | Structure of DHODH in Complex with Ligand 17 | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of Alternative Binding Poses through Fragment-Based Identification of DHODH Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8VHH
 
 | | Engineered holo tryptophan synthase (Tm9D8*) derived from T. maritima TrpB | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Porter, N.J, Johnston, K.E, Almhjell, P.J, Arnold, F.H. | | Deposit date: | 2024-01-02 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A combinatorially complete epistatic fitness landscape in an enzyme active site.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VHE
 
 | | Crystal Structure of Human IDH1 R132Q in Complex with NADPH-TCEP Adduct | | Descriptor: | 3,3',3''-({(4R)-1-[(2R,3R,4S,5R)-5-({[(S)-{[(S)-{[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-3,4-dihydroxyoxolan-2-yl]-3-carbamoyl-1,4-dihydropyridin-4-yl}-lambda~5~-phosphanetriyl)tripropanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
8VHD
 
 | | Crystal Structure of Human IDH1 R132Q in complex with NADPH and Isocitrate | | Descriptor: | CALCIUM ION, GLYCEROL, IODIDE ION, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
8VHC
 
 | | Crystal Structure of Human IDH1 R132Q in complex with NADPH | | Descriptor: | GLYCEROL, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
8VHB
 
 | | Crystal Structure of Human IDH1 R132Q in complex with NADPH and Alpha-Ketoglutarate | | Descriptor: | (3~{S})-3-[(4~{S})-3-aminocarbonyl-1-[(2~{R},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]-4~{H}-pyridin-4-yl]-2-oxidanylidene-pentanedioic acid, 2-OXOGLUTARIC ACID, CALCIUM ION, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
