1SDZ
 
 | | Crystal structure of DIAP1 BIR1 bound to a Reaper peptide | | Descriptor: | Apoptosis 1 inhibitor, Reaper, ZINC ION | | Authors: | Yan, N, Wu, J.W, Shi, Y. | | Deposit date: | 2004-02-15 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3LNG
 
 | |
1E2S
 
 | | Crystal structure of an Arylsulfatase A mutant C69A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION, ... | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-05-24 | | Release date: | 2000-12-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
J. Mol. Biol., 305, 2001
|
|
1E3C
 
 | | Crystal structure of an Arylsulfatase A mutant C69S soaked in synthetic substrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-06-13 | | Release date: | 2001-03-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
J. Mol. Biol., 305, 2001
|
|
1E3Z
 
 | | Acarbose complex of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.93A | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1UWH
 
 | | The complex of wild type B-RAF and BAY439006. | | Descriptor: | 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, B-RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE, CHLORIDE ION | | Authors: | Barford, D, Roe, S.M, Wan, P.T.C, Cancer Genome Project | | Deposit date: | 2004-02-05 | | Release date: | 2004-03-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Mechanism of Activation of the Raf-Erk Signaling Pathway by Oncogenic Mutations of B-Raf
Cell(Cambridge,Mass.), 116, 2004
|
|
3LQV
 
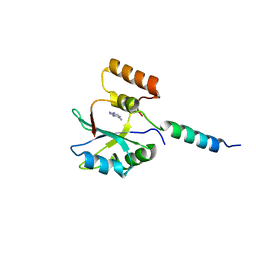 | | Branch Recognition by SF3b14 | | Descriptor: | ADENINE, Pre-mRNA branch site protein p14, Splicing factor 3B subunit 1 | | Authors: | Schellenberg, M.J, MacMillan, A.M. | | Deposit date: | 2010-02-10 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Branch Recognition by SF3b14
Rna, 17, 2011
|
|
1BF5
 
 | | TYROSINE PHOSPHORYLATED STAT-1/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*G P*C)-3'), DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*CP*T P*G)-3'), SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION 1-ALPHA/BETA | | Authors: | Kuriyan, J, Zhao, Y, Chen, X, Vinkemeier, U, Jeruzalmi, D, Darnell Jr, J.E. | | Deposit date: | 1998-05-27 | | Release date: | 1998-08-12 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA.
Cell(Cambridge,Mass.), 93, 1998
|
|
1UWJ
 
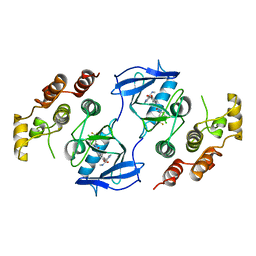 | | The complex of mutant V599E B-RAF and BAY439006. | | Descriptor: | 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, B-RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Barford, D, Roe, S.M, Wan, P.T.C, Cancer Genome Project | | Deposit date: | 2004-02-05 | | Release date: | 2004-03-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Mechanism of Activation of the Raf-Erk Signaling Pathway by Oncogenic Mutations of B-Raf
Cell(Cambridge,Mass.), 116, 2004
|
|
1UW4
 
 | |
1BIF
 
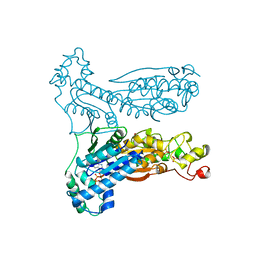 | | 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE BIFUNCTIONAL ENZYME COMPLEXED WITH ATP-G-S AND PHOSPHATE | | Descriptor: | 6-PHOSPHOFRUCTO-2-KINASE/ FRUCTOSE-2,6-BISPHOSPHATASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Hasemann, C.A, Deisenhofer, J. | | Deposit date: | 1996-11-08 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the bifunctional enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase reveals distinct domain homologies.
Structure, 4, 1996
|
|
1SGT
 
 | |
1BGF
 
 | |
1E2V
 
 | | N153Q mutant of cytochrome f from Chlamydomonas reinhardtii | | Descriptor: | ACETATE ION, CYTOCHROME F, HEME C | | Authors: | Sainz, G, Carrell, C.J, Ponamarev, M.V, Soriano, G.M, Cramer, W.A, Smith, J.L. | | Deposit date: | 2000-05-29 | | Release date: | 2000-08-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Interruption of the Internal Water Chain of Cytochrome F Impairs Photosynthetic Function
Biochemistry, 39, 2000
|
|
1DRB
 
 | |
1E2Z
 
 | | Q158L mutant of cytochrome f from Chlamydomonas reinhardtii | | Descriptor: | CYTOCHROME F, HEME C | | Authors: | Sainz, G, Carrell, C.J, Ponamarev, M.V, Soriano, G.M, Cramer, W.A, Smith, J.L. | | Deposit date: | 2000-05-30 | | Release date: | 2000-08-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interruption of the Internal Water Chain of Cytochrome F Impairs Photosynthetic Function
Biochemistry, 39, 2000
|
|
1DXO
 
 | | Crystal structure of human NAD[P]H-QUINONE oxidoreductase CO with 2,3,5,6,tetramethyl-P-benzoquinone (duroquinone) at 2.5 Angstrom resolution | | Descriptor: | DUROQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2000-01-12 | | Release date: | 2000-04-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Recombinant Mouse and Human Nad(P)H:Quinone Oxidoreductases:Species Comparison and Structural Changes with Substrate Binding and Release
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DTW
 
 | | HUMAN BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE | | Descriptor: | BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE ALPHA SUBUNIT, BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE BETA SUBUNIT, MAGNESIUM ION, ... | | Authors: | AEvarsson, A, Chuang, J.L, Wynn, R.M, Turley, S, Chuang, D.T, Hol, W.G.J. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human branched-chain alpha-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease.
Structure Fold.Des., 8, 2000
|
|
1E1Z
 
 | | Crystal structure of an Arylsulfatase A mutant C69S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION | | Authors: | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | Deposit date: | 2000-05-12 | | Release date: | 2001-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
J. Mol. Biol., 305, 2001
|
|
1SP2
 
 | |
1DWX
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and tetrahydrobiopterin | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
1SP1
 
 | |
1SBP
 
 | |
1CNN
 
 | | OMEGA-CONOTOXIN MVIIC FROM CONUS MAGUS | | Descriptor: | OMEGA-CONOTOXIN MVIIC | | Authors: | Nielsen, K.J, Adams, D, Thomas, L, Bond, T, Alewood, P.F, Craik, D.J, Lewis, R.J. | | Deposit date: | 1999-05-20 | | Release date: | 2000-05-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-activity relationships of omega-conotoxins MVIIA, MVIIC and 14 loop splice hybrids at N and P/Q-type calcium channels.
J.Mol.Biol., 289, 1999
|
|
1D8D
 
 | | CO-CRYSTAL STRUCTURE OF RAT PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH A K-RAS4B PEPTIDE SUBSTRATE AND FPP ANALOG AT 2.0A RESOLUTION | | Descriptor: | ACETATE ION, K-RAS4B PEPTIDE SUBSTRATE, ZINC ION, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 1999-10-22 | | Release date: | 2000-02-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The basis for K-Ras4B binding specificity to protein farnesyltransferase revealed by 2 A resolution ternary complex structures.
Structure Fold.Des., 8, 2000
|
|
