3LQ0
 
 | | Zymogen structure of crayfish astacin metallopeptidase | | Descriptor: | GLYCEROL, ProAstacin, SULFATE ION, ... | | Authors: | Guevara, T, Yiallouros, I, Kappelhoff, R, Bissdorf, S, Stocker, W, Gomis-Ruth, F.X. | | Deposit date: | 2010-02-08 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Proenzyme structure and activation of astacin metallopeptidase
J.Biol.Chem., 285, 2010
|
|
1UW0
 
 | |
5VOM
 
 | | Benzopiperazine BET bromodomain inhibitor in complex with BD1 of Brd4 | | Descriptor: | 3-[(2S)-1-acetyl-4-(furan-2-carbonyl)-2-methyl-1,2,3,4-tetrahydroquinoxalin-6-yl]-N-methylbenzamide, Bromodomain-containing protein 4 | | Authors: | Toms, A.V, Herbertz, T. | | Deposit date: | 2017-05-03 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Design and Optimization of Benzopiperazines as Potent Inhibitors of BET Bromodomains.
ACS Med Chem Lett, 8, 2017
|
|
5W49
 
 | | The crystal structure of human S-adenosylhomocysteine hydrolase (AHCY) bound to oxadiazole inhibitor | | Descriptor: | (4-amino-1,2,5-oxadiazol-3-yl)[(3R)-3-{4-[(3-methoxyphenyl)amino]-6-methylpyridin-2-yl}pyrrolidin-1-yl]methanone, 1,2-ETHANEDIOL, Adenosylhomocysteinase, ... | | Authors: | Dougan, D.R, Lawson, J.D, Lane, W. | | Deposit date: | 2017-06-09 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of AHCY inhibitors using novel high-throughput mass spectrometry.
Biochem. Biophys. Res. Commun., 491, 2017
|
|
5VQ1
 
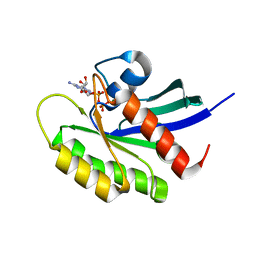 | | Crystal structure of human KRAS Q61A mutant in complex with GDP | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, S, Long, B, Boris, G, Ni, S, Kennedy, M.A. | | Deposit date: | 2017-05-07 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural insight into the rearrangement of the switch I region in GTP-bound G12A K-Ras.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1UYA
 
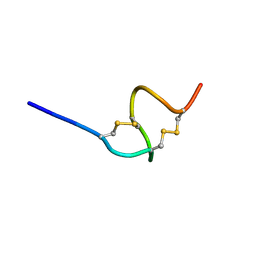 | | THE SOLUTION STRUCTURE OF THE A-FORM OF UROGUANYLIN-16 NMR, 10 STRUCTURES | | Descriptor: | UROGUANYLIN-16, ISOMER A | | Authors: | Marx, U.C, Adermann, K, Forssmann, W.-G, Roesch, P. | | Deposit date: | 1997-09-11 | | Release date: | 1998-03-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | One peptide, two topologies: structure and interconversion dynamics of human uroguanylin isomers.
J.Pept.Res., 52, 1998
|
|
3L82
 
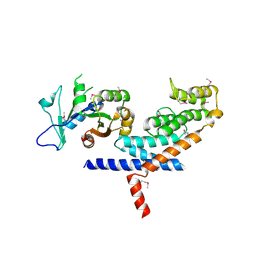 | | X-ray Crystal structure of TRF1 and Fbx4 complex | | Descriptor: | F-box only protein 4, Telomeric repeat-binding factor 1 | | Authors: | Zeng, Z.X, Wang, W, Yang, Y.T, Chen, Y, Yang, X.M, Diehl, J.A, Liu, X.D, Lei, M. | | Deposit date: | 2009-12-29 | | Release date: | 2010-03-09 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Selective Ubiquitination of TRF1 by SCF(Fbx4)
Dev.Cell, 18, 2010
|
|
1UYB
 
 | | THE SOLUTION STRUCTURE OF THE B-FORM OF UROGUANYLIN-16 NMR, 10 STRUCTURES | | Descriptor: | UROGUANYLIN-16, ISOMER B | | Authors: | Marx, U.C, Adermann, K, Forssmann, W.-G, Roesch, P. | | Deposit date: | 1997-09-11 | | Release date: | 1998-03-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | One peptide, two topologies: structure and interconversion dynamics of human uroguanylin isomers.
J.Pept.Res., 52, 1998
|
|
1E21
 
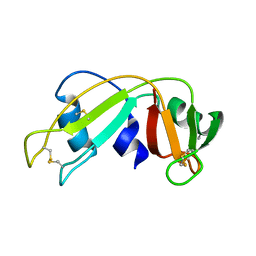 | | Ribonuclease 1 des1-7 Crystal Structure at 1.9A | | Descriptor: | RIBONUCLEASE 1 | | Authors: | Pous, J, Mallorqui-Fernandez, G, Peracaula, R, Terzyan, S.S, Futami, J, Tada, H, Yamada, H, Seno, M, De Llorens, R, Gomis-Ruth, F.X, Coll, M. | | Deposit date: | 2000-05-15 | | Release date: | 2001-05-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Crystal Structure of Human Rnase 1Dn7 at 1.9A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1E2W
 
 | | N168F mutant of cytochrome f from Chlamydomonas reinhardtii | | Descriptor: | CYTOCHROME F, HEME C | | Authors: | Sainz, G, Carrell, C.J, Ponamarev, M.V, Soriano, G.M, Cramer, W.A, Smith, J.L. | | Deposit date: | 2000-05-30 | | Release date: | 2000-08-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interruption of the Internal Water Chain of Cytochrome F Impairs Photosynthetic Function
Biochemistry, 39, 2000
|
|
5VRY
 
 | | Human DNA polymerase beta 8-oxoG:dC extension with dTTP after 20 s | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*(8OG)P*GP*CP*GP*CP*AP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*TP*GP*CP*GP*CP*CP*T)-3'), ... | | Authors: | Reed, A.J, Suo, Z. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Time-Dependent Extension from an 8-Oxoguanine Lesion by Human DNA Polymerase Beta.
J. Am. Chem. Soc., 139, 2017
|
|
5VS3
 
 | | Human DNA polymerase beta 8-oxoG:dA extension with dTTP after 90 s | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*(8OG)P*GP*CP*GP*CP*AP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*TP*GP*CP*GP*CP*AP*T)-3'), ... | | Authors: | Reed, A.J, Suo, Z. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Time-Dependent Extension from an 8-Oxoguanine Lesion by Human DNA Polymerase Beta.
J. Am. Chem. Soc., 139, 2017
|
|
5VO4
 
 | |
1S05
 
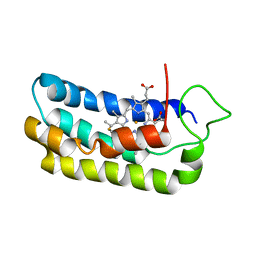 | | NMR-validated structural model for oxidized R.palustris cytochrome c556 | | Descriptor: | Cytochrome c-556, HEME C | | Authors: | Bertini, I, Faraone-Mennella, J, Gray, H.B, Luchinat, C, Parigi, G, Winkler, J.R. | | Deposit date: | 2003-12-30 | | Release date: | 2004-01-20 | | Last modified: | 2021-03-03 | | Method: | SOLUTION NMR | | Cite: | NMR-validated structural model for oxidized Rhodopseudomonas palustris cytochrome c(556).
J.Biol.Inorg.Chem., 9, 2004
|
|
5W2C
 
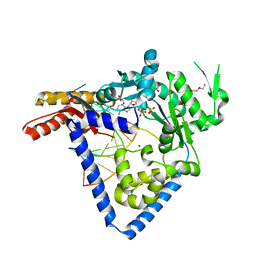 | |
1DWW
 
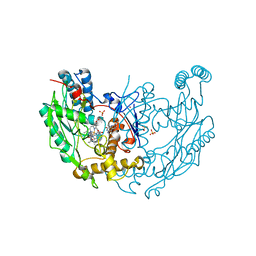 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and dihydrobiopterin | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
5W5I
 
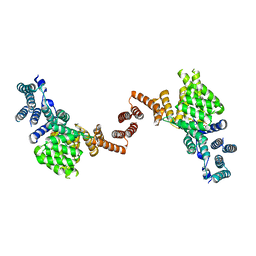 | | Human IFIT1 dimer with PPP-AAAA | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 1, RNA (5'-D(*(ATP))-R(P*AP*AP*A)-3') | | Authors: | Abbas, Y.M, Martinez-Montero, S, Damha, M.J, Nagar, B. | | Deposit date: | 2017-06-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into IFIT1 dimerization and conformational changes associated with mRNA binding
To Be Published
|
|
1S4Y
 
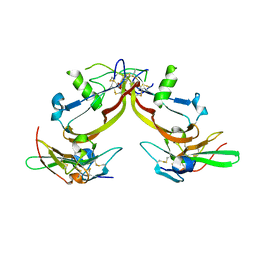 | | Crystal structure of the activin/actrIIb extracellular domain | | Descriptor: | Activin receptor type IIB precursor, Inhibin beta A chain | | Authors: | Greenwald, J, Vega, M.E, Allendorph, G.P, Fischer, W.H, Vale, W, Choe, S, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-01-19 | | Release date: | 2004-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Flexible Activin Explains the Membrane-Dependent Cooperative Assembly of TGF-beta Family Receptors.
Mol.Cell, 15, 2004
|
|
5VSR
 
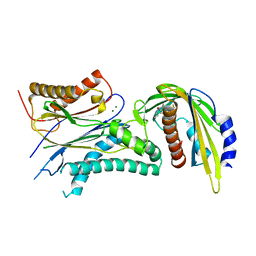 | | ABA-mimicking ligand AMF4 in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)-1-(2,3,5,6-tetrafluoro-4-methylphenyl)methanesulfonamide, ... | | Authors: | Cao, M.-J, Zhang, Y.-L, Liu, X, Huang, H, Zhou, X.E, Wang, W.-L, Zeng, A, Zhao, C.-Z, Si, T, Du, J.-M, Wu, W.-W, Wang, F.-X, Xu, H.X, Zhu, J.-K. | | Deposit date: | 2017-05-12 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.618 Å) | | Cite: | Combining chemical and genetic approaches to increase drought resistance in plants.
Nat Commun, 8, 2017
|
|
1SC3
 
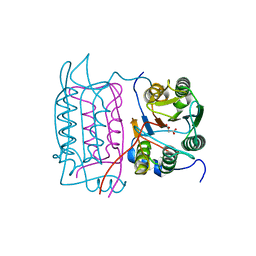 | | Crystal structure of the human caspase-1 C285A mutant in complex with malonate | | Descriptor: | Interleukin-1 beta convertase, MALONATE ION | | Authors: | Romanowski, M.J, Scheer, J.M, O'Brien, T, McDowell, R.S. | | Deposit date: | 2004-02-11 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of a ligand-free and malonate-bound human caspase-1: implications for the mechanism of substrate binding.
Structure, 12, 2004
|
|
1SH5
 
 | |
5VQ0
 
 | | Crystal structure of human KRAS G12A mutant in complex with GDP (EDTA soaked) | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Xu, S, Long, B, Boris, G, Ni, S, Kennedy, M.A. | | Deposit date: | 2017-05-06 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into the rearrangement of the switch I region in GTP-bound G12A K-Ras.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5VQ6
 
 | | Crystal structure of human WT-KRAS in complex with GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase KRas, MAGNESIUM ION | | Authors: | Xu, S, Long, B, Boris, G, Ni, S, Kennedy, M.A. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insight into the rearrangement of the switch I region in GTP-bound G12A K-Ras.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3LHA
 
 | |
1SA4
 
 | | human protein farnesyltransferase complexed with FPP and R115777 | | Descriptor: | 6-[(S)-AMINO(4-CHLOROPHENYL)(1-METHYL-1H-IMIDAZOL-5-YL)METHYL]-4-(3-CHLOROPHENYL)-1-METHYLQUINOLIN-2(1H)-ONE, FARNESYL DIPHOSPHATE, Protein farnesyltransferase beta subunit, ... | | Authors: | Reid, T.S, Beese, L.S. | | Deposit date: | 2004-02-06 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Anticancer Clinical Candidates R115777 (Tipifarnib) and BMS-214662 Complexed with Protein Farnesyltransferase Suggest a Mechanism of FTI Selectivity.
Biochemistry, 43, 2004
|
|
