6E7X
 
 | |
6E7U
 
 | |
7V8S
 
 | | Crystal structure of cyclohexanone monooxygenase from T. municipale mutant L437T complexed with NADP+ and FAD in space group of P1211 | | Descriptor: | Cyclohexanone Monooxygenase from Thermocrispum municipale, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Li, T, Li, G.Y, Yin, H. | | Deposit date: | 2021-08-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Biocatalytic Baeyer-Villiger Reactions: Uncovering the Source of Regioselectivity at Each Evolutionary Stage of a Mutant with Scrutiny of Fleeting Chiral Intermediates.
Acs Catalysis, 12, 2022
|
|
7V8R
 
 | | Crystal structure of cyclohexanone monooxygenase from T. municipale mutant L437T complexed with NADP+ and FAD in space group of C2221 | | Descriptor: | 1,2-ETHANEDIOL, Cyclohexanone Monooxygenase from Thermocrispum municipale, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Li, T, Li, G.Y, Yin, H. | | Deposit date: | 2021-08-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Biocatalytic Baeyer-Villiger Reactions: Uncovering the Source of Regioselectivity at Each Evolutionary Stage of a Mutant with Scrutiny of Fleeting Chiral Intermediates.
Acs Catalysis, 12, 2022
|
|
6EJ7
 
 | |
7KGT
 
 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N226-234 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A*02:01 molecule.
Iscience, 24, 2021
|
|
6EFD
 
 | | Hsa Siglec and Unique domains in complex with the sialyl T antigen trisaccharide | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose, SODIUM ION, Streptococcal hemagglutinin | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
7VMF
 
 | |
7KYR
 
 | |
7VH4
 
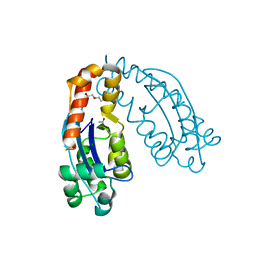 | | Crystal structure of oligoribonuclease of Escherichia coli | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Badhwar, P, Taneja, B. | | Deposit date: | 2021-09-20 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of a mycobacterial oligoribonuclease reveals a unique C-terminal tail that stabilizes the homodimer.
J.Biol.Chem., 298, 2022
|
|
7L6C
 
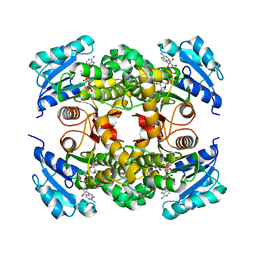 | |
7KUT
 
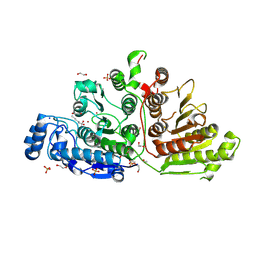 | | Crystal Structure of Danio rerio Histone Deacetylase 10 H137A Mutant in Complex with N-Acetylputrescine (Tetrahedral Intermediate) | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4-aminobutyl)amino]ethane-1,1-diol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Herbst-Gervasoni, C.J, Christianson, D.W. | | Deposit date: | 2020-11-25 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray Crystallographic Snapshots of Substrate Binding in the Active Site of Histone Deacetylase 10.
Biochemistry, 60, 2021
|
|
7VO7
 
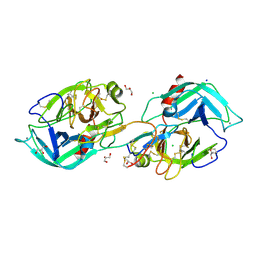 | | Crystal structure of trypsin in complex with Lima bean trypsin inhibitor at 2.25A resolution. | | Descriptor: | Bowman-Birk type proteinase inhibitor, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ahmad, M.S, Akbar, Z, Choudhary, M.I. | | Deposit date: | 2021-10-12 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insight into the structural basis of the dual inhibitory mode of Lima bean (Phaseolus lunatus) serine protease inhibitor.
Proteins, 91, 2023
|
|
7KIA
 
 | | Crystal structure of FGFR2 kinase domain gatekeeper mutant V564F in complex with covalent compound 19 | | Descriptor: | 1-[4-(4-{4-(4-methylpiperazin-1-yl)-6-[(3-methyl-1H-pyrazol-5-yl)amino]pyrimidin-2-yl}phenyl)piperidin-1-yl]prop-2-en-1-one, CITRATE ANION, Fibroblast growth factor receptor 2, ... | | Authors: | Ke, J, Wibowo, A.S, Carter, J.J, Larsen, N.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of Aminopyrazole Derivatives as Potent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR2 and 3.
Acs Med.Chem.Lett., 12, 2021
|
|
7KV2
 
 | | Surface glycan-binding protein A from Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, RagB/SusD family nutrient uptake outer membrane protein, ... | | Authors: | Tamura, K, Brumer, H, Van Petegem, F. | | Deposit date: | 2020-11-26 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Distinct protein architectures mediate species-specific beta-glucan binding and metabolism in the human gut microbiota.
J.Biol.Chem., 296, 2021
|
|
7VWD
 
 | | Crystal Structure of the Y53F/N55A mutant of LEH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2021-11-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7KZA
 
 | |
4MLM
 
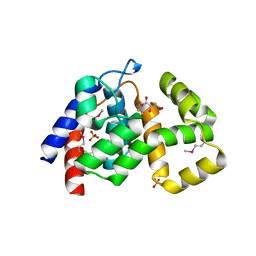 | | Crystal Structure of PhnZ from uncultured bacterium HF130_AEPn_1 | | Descriptor: | FE (III) ION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | van Staalduinen, L.M, McSorley, F.R, Zechel, D.L, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of PhnZ in complex with substrate reveals a di-iron oxygenase mechanism for catabolism of organophosphonates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6F0R
 
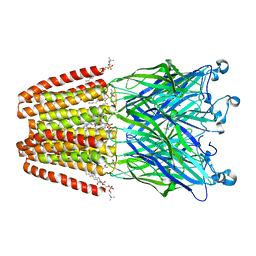 | | GLIC mutant E82Q | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Full mutational mapping of titratable residues helps to identify proton-sensors involved in the control of channel gating in the Gloeobacter violaceus pentameric ligand-gated ion channel.
PLoS Biol., 15, 2017
|
|
7L7X
 
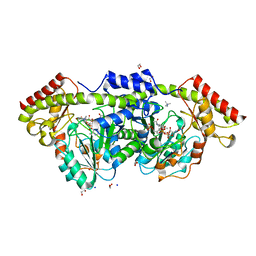 | | X-ray structure of the Pcryo_0638 aminotransferase from Psychrobacter cryohalolentis | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, DegT/DnrJ/EryC1/StrS aminotransferase, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
4MMF
 
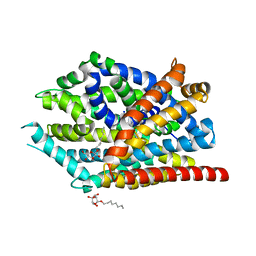 | | Crystal structure of LeuBAT (delta5 mutant) in complex with mazindol | | Descriptor: | (5R)-5-(4-chlorophenyl)-2,5-dihydro-3H-imidazo[2,1-a]isoindol-5-ol, SODIUM ION, Transporter, ... | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
7L1A
 
 | | Human Methionine Adenosyltransferase 2A bound to Methylthioadenosine and inhibitor, di-imido triphosphate (PNPNP) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALANINE, MAGNESIUM ION, ... | | Authors: | Niland, C.N, Fedorov, E, Schramm, V.L, Ghosh, A. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanism and Inhibition of Human Methionine Adenosyltransferase 2A.
Biochemistry, 60, 2021
|
|
6EVV
 
 | | X-ray structure of the complex between human alpha thrombin and NU172, a duplex/quadruplex 26-mer DNA aptamer, in the presence of potassium ions. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Troisi, R, Russo Krauss, I, Sica, F. | | Deposit date: | 2017-11-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer.
Nucleic Acids Res., 46, 2018
|
|
4MS2
 
 | | Structural basis of Ca2+ selectivity of a voltage-gated calcium channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-18 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
4MTF
 
 | | Structural Basis of Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-19 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
