4AMK
 
 | |
3Q0J
 
 | |
4QKW
 
 | | Crystal structure of the zebrafish cavin4a HR1 domain | | Descriptor: | CHLORIDE ION, Muscle-related coiled-coil protein | | Authors: | Kovtun, O, Tillu, V, Parton, R.G, Collins, B.M. | | Deposit date: | 2014-06-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the organization of the cavin membrane coat complex
Dev.Cell, 31, 2014
|
|
2W2I
 
 | | Crystal structure of the human 2-oxoglutarate oxygenase LOC390245 | | Descriptor: | 2-OXOGLUTARATE OXYGENASE, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Yue, W.W, Ng, S, Shafqat, N, Ugochukwu, E, McDonough, M, Pike, A.C.W, Filippakopoulos, P, von Delft, F, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Schofield, C, Oppermann, U. | | Deposit date: | 2008-10-31 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Evolutionary Basis for the Dual Substrate Selectivity of Human Kdm4 Histone Demethylase Family.
J.Biol.Chem., 286, 2011
|
|
4RFE
 
 | | Crystal structure of ADCC-potent ANTI-HIV-1 Rhesus macaque antibody JR4 Fab | | Descriptor: | CHLORIDE ION, Fab heavy chain of ADCC-potent anti-HIV-1 antibody JR4, Fab light chain of ADCC-potent anti-HIV-1 antibody JR4, ... | | Authors: | Wu, X, Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
1S79
 
 | | Solution structure of the central RRM of human La protein | | Descriptor: | Lupus La protein | | Authors: | Alfano, C, Sanfelice, D, Babon, J, Kelly, G, Jacks, A, Curry, S, Conte, M.R. | | Deposit date: | 2004-01-29 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2XQ4
 
 | | Pentameric ligand gated ion channel GLIC in complex with tetramethylarsonium (TMAs) | | Descriptor: | ARSENIC, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
4OCI
 
 | | Crystal Structure of Calcium Binding Protein-5 from Entamoeba histolytica and its involvement in initiation of phagocytosis of human erythrocytes | | Descriptor: | ACETATE ION, CALCIUM ION, Calmodulin, ... | | Authors: | Kumar, S, Manjasetty, A.B, Zaidi, R, Gourinath, S. | | Deposit date: | 2014-01-09 | | Release date: | 2014-12-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Crystal Structure of Calcium Binding Protein-5 from Entamoeba histolytica and Its Involvement in Initiation of Phagocytosis of Human Erythrocytes.
Plos Pathog., 10, 2014
|
|
3PCG
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH THE INHIBITOR 4-HYDROXYPHENYLACETATE | | Descriptor: | 4-HYDROXYPHENYLACETATE, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Elango, N, Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-04-29 | | Release date: | 1998-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
3PCE
 
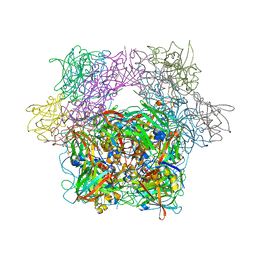 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3-HYDROXYPHENYLACETATE | | Descriptor: | 3-HYDROXYPHENYLACETATE, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Elango, N, Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-04-29 | | Release date: | 1998-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
1SE9
 
 | |
2Y41
 
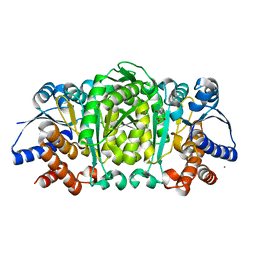 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus - complex with IPM and MN | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, 3-ISOPROPYLMALIC ACID, MANGANESE (II) ION | | Authors: | Graczer, E, merlin, A, Singh, R.K, Manikandan, K, Zavodsky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Atomic Level Description of the Domain Closure in a Dimeric Enzyme: Thermus Thermophilus 3-Isopropylmalate Dehydrogenase.
Mol.Biosyst., 7, 2011
|
|
2Y3Z
 
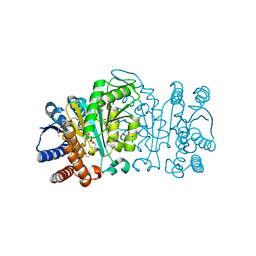 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus - apo enzyme | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-ISOPROPYLMALATE DEHYDROGENASE, GLYCEROL, ... | | Authors: | Graczer, E, merlin, A, Singh, R.K, Manikandan, K, Zavodsky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Atomic Level Description of the Domain Closure in a Dimeric Enzyme: Thermus Thermophilus 3-Isopropylmalate Dehydrogenase.
Mol.Biosyst., 7, 2011
|
|
2Y40
 
 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus - complex with Mn | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, MANGANESE (II) ION | | Authors: | Graczer, E, merlin, A, Singh, R.K, Manikandan, K, Zavodsky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Level Description of the Domain Closure in a Dimeric Enzyme: Thermus Thermophilus 3-Isopropylmalate Dehydrogenase.
Mol.Biosyst., 7, 2011
|
|
4PE2
 
 | | MBP PilA1 CD160 | | Descriptor: | MALONATE ION, Maltose ABC transporter periplasmic protein,Prepilin-type N-terminal cleavage/methylation domain protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Piepenbrink, K.H, Sundberg, E.J. | | Deposit date: | 2014-04-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | Structural and Evolutionary Analyses Show Unique Stabilization Strategies in the Type IV Pili of Clostridium difficile.
Structure, 23, 2015
|
|
4IDY
 
 | | Mycobacterium Tuberculosis Methionine aminopeptidase Type 1c in complex with 2-hydroxyethyl disulfide | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Methionine aminopeptidase 2, POTASSIUM ION | | Authors: | Reddi, R, Gumpena, R, Kishor, C, Addlagatta, A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective targeting of the conserved active site cysteine of Mycobacterium tuberculosis methionine aminopeptidase with electrophilic reagents
Febs J., 281, 2014
|
|
3NTN
 
 | | Crystal Structure of UspA1 head and neck domain from Moraxella catarrhalis | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Conners, R, Zaccai, N, Agnew, C, Burton, N, Brady, R.L. | | Deposit date: | 2010-07-05 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Correlation of in situ mechanosensitive responses of the Moraxella catarrhalis adhesin UspA1 with fibronectin and receptor CEACAM1 binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NUG
 
 | |
4S2E
 
 | |
4BCP
 
 | | Structure of CDK2 in complex with cyclin A and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 2-[[3-(1,4-diazepan-1-yl)phenyl]amino]-4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]pyrimidine-5-carbonitrile, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2, ... | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Substituted 4-(Thiazol-5-Yl)-2-(Phenylamino)Pyrimidines are Highly Active Cdk9 Inhibitors: Synthesis, X-Ray Crystal Structure, Sar and Anti-Cancer Activities.
J.Med.Chem., 56, 2013
|
|
4IF7
 
 | | Mycobacterium Tuberculosis Methionine aminopeptidase Type 1c in complex with homocysteine-methyl disulfide | | Descriptor: | (2S)-2-amino-4-(methyldisulfanyl)butanoic acid, COBALT (II) ION, Methionine aminopeptidase 2 | | Authors: | Reddi, R, Gumpena, R, Kishor, C, Addlagatta, A. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective targeting of the conserved active site cysteine of Mycobacterium tuberculosis methionine aminopeptidase with electrophilic reagents
Febs J., 281, 2014
|
|
4JY1
 
 | | CRYSTAL STRUCTURE OF HCV NS5B POLYMERASE IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 3-{ISOPROPYL[(TRANS-4-METHYLCYCLOHEXYL)CARBONYL]AMINO}-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, Genome polyprotein | | Authors: | Coulombe, R. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinguishing drug binding pockets on proteins by complementary biophysical and biological methods
To be Published
|
|
4TPM
 
 | |
4JE7
 
 | | Crystal structure of a human-like mitochondrial peptide deformylase in complex with actinonin | | Descriptor: | ACTINONIN, Peptide deformylase 1A, chloroplastic/mitochondrial, ... | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2WVM
 
 | | H309A mutant of Mannosyl-3-phosphoglycerate synthase from Thermus thermophilus HB27 in complex with GDP-alpha-D-Mannose and Mg(II) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, MANNOSYL-3-PHOSPHOGLYCERATE SYNTHASE, ... | | Authors: | Goncalves, S, Borges, N, Esteves, A.M, Victor, B, Soares, C.M, Santos, H, Matias, P.M. | | Deposit date: | 2009-10-19 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.977 Å) | | Cite: | Structural Analysis of Thermus Thermophilus Hb27 Mannosyl-3-Phosphoglycerate Synthase Provides Evidence for a Second Catalytic Metal Ion and New Insight Into the Retaining Mechanism of Glycosyltransferases.
J.Biol.Chem., 285, 2010
|
|
