4KZ8
 
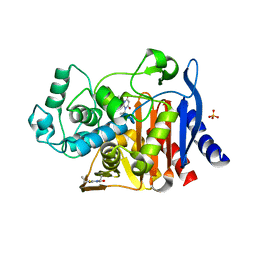 | | Crystal structure of AmpC beta-lactamase in complex with fragment 20 (1,3-diethyl-2-thioxodihydropyrimidine-4,6(1H,5H)-dione) | | Descriptor: | 1,3-diethyl-2-thioxodihydropyrimidine-4,6(1H,5H)-dione, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ4
 
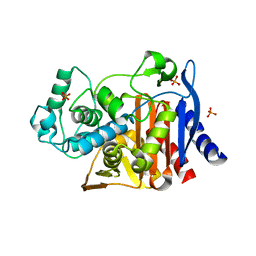 | | Crystal structure of AmpC beta-lactamase in complex with fragment 60 (2-[(propylsulfonyl)amino]benzoic acid) | | Descriptor: | 2-[(propylsulfonyl)amino]benzoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
2J8T
 
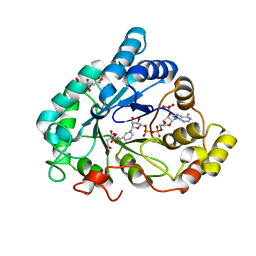 | | Human aldose reductase in complex with NADP and citrate at 0.82 angstrom | | Descriptor: | ALDO-KETO REDUCTASE FAMILY 1, MEMBER B1, CITRATE ANION, ... | | Authors: | Biadene, M, Hazemann, I, Cousido, A, Ginell, S, Sheldrick, G.M, Podjarny, A, Schneider, T.R. | | Deposit date: | 2006-10-27 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.82 Å) | | Cite: | The Atomic Resolution Structure of Human Aldose Reductase Reveals that Rearrangement of a Bound Ligand Allows the Opening of the Safety-Belt Loop.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4B15
 
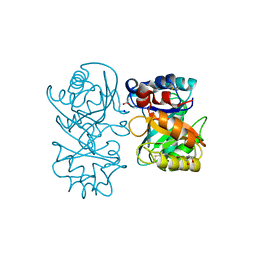 | | crystal structure of tamarind chitinase like lectin (TCLL) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Patil, D.N, Kumar, P. | | Deposit date: | 2012-07-06 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Investigation of a Novel N-Acetyl Glucosamine Binding Chi-Lectin which Reveals Evolutionary Relationship with Class III Chitinases.
Plos One, 8, 2013
|
|
5CNB
 
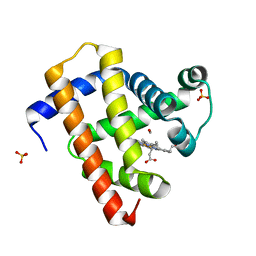 | | Ultrafast dynamics in myoglobin: 0.5 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
5CNG
 
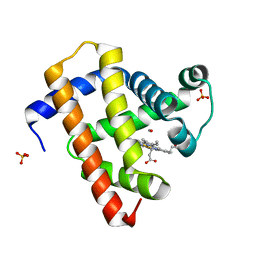 | | ultrafast dynamics in myoglobin: 150 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
5CN9
 
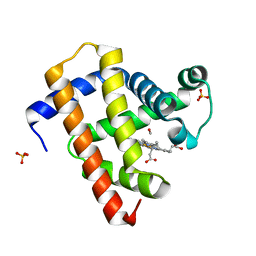 | | Ultrafast dynamics in myoglobin: 0.4 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
6AZJ
 
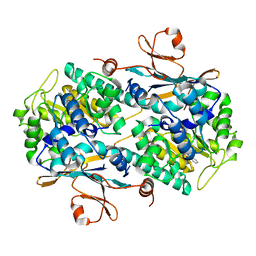 | | Crystal Structure of human NAMPT in complex with NVP-LQN520 | | Descriptor: | (1S,2S)-N-{4-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]phenyl}-2-(pyridin-3-yl)cyclopropane-1-carboxamide, Nicotinamide phosphoribosyltransferase | | Authors: | Weihofen, W.A, Thigale, S. | | Deposit date: | 2017-09-11 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification and structure based design of cellularly active cyclo-propyl carboxamide Nicotinamide phosphoribosyltransferase (NAMPT) inhibitors
To Be Published
|
|
4B16
 
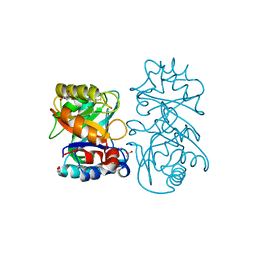 | |
4B14
 
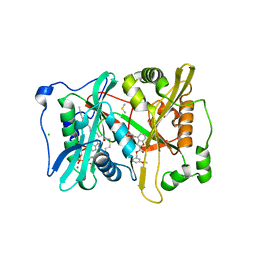 | | Plasmodium vivax N-myristoyltransferase with a bound benzofuran inhibitor (compound 26) | | Descriptor: | 2-oxopentadecyl-CoA, 3-methoxybenzyl 3-methyl-4-(piperidin-4-yloxy)-1-benzofuran-2-carboxylate, CHLORIDE ION, ... | | Authors: | Yu, Z, Brannigan, J.A, Moss, D.K, Brzozowski, A.M, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2012-07-06 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design and Synthesis of Inhibitors of Plasmodium Falciparum N-Myristoyltransferase, a Promising Target for Anti-Malarial Drug Discovery.
J.Med.Chem., 55, 2012
|
|
7LA5
 
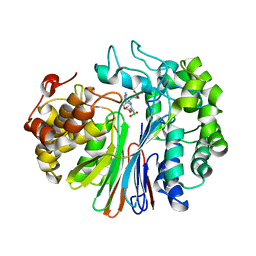 | | Structure of human GGT1 in complex with Lnt1-172 compound. | | Descriptor: | (2R)-4-borono-2-{[(1H-imidazol-4-yl)methyl]amino}butanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Terzyan, S.S, Hanigan, M, Nguen, L. | | Deposit date: | 2021-01-05 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of human GGT1 in complex with Lnt1-172 compound.
To Be Published
|
|
2OGW
 
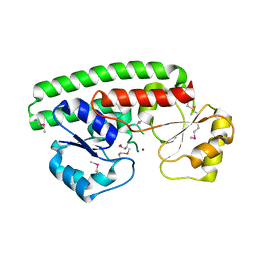 | | Structure of ABC type zinc transporter from E. coli | | Descriptor: | High-affinity zinc uptake system protein znuA precursor, ZINC ION | | Authors: | Sharma, A, Yogavel, M. | | Deposit date: | 2007-01-09 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Analysis of ABC-family Periplasmic Zinc Binding Protein Provides New Insights Into Mechanism of Ligand Uptake and Release
J.Mol.Biol., 367, 2007
|
|
7XK8
 
 | | Cryo-EM structure of the Neuromedin U receptor 2 (NMUR2) in complex with G Protein and its endogeneous Peptide-Agonist NMU25 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, W, Wenru, Z, Mu, W, Minmin, L, Shutian, C, Tingting, T, Gisela, S, Holger, W, Albert, B, Cuiying, Y, Xiaojing, C, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and activation of neuromedin U receptor 2.
Nat Commun, 13, 2022
|
|
4LMU
 
 | | Crystal structure of Pim1 in complex with the inhibitor Quercetin (resulting from displacement of SKF86002) | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4R88
 
 | | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-fluorocytosine | | Descriptor: | 1,2-ETHANEDIOL, 5-fluorocytosine, ACETIC ACID, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-08-29 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-fluorocytosine
To be Published
|
|
6CVH
 
 | | Identification and biological evaluation of thiazole-based inverse agonists of RORgt | | Descriptor: | Nuclear receptor ROR-gamma, trans-3-({4-(cyclohexylmethyl)-5-[3-(1-methylcyclopropyl)-5-{[(2R)-1,1,1-trifluoropropan-2-yl]carbamoyl}phenyl]-1,3-thiazole-2-carbonyl}amino)cyclobutane-1-carboxylic acid | | Authors: | Spurlino, J, Milligan, C. | | Deposit date: | 2018-03-28 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Identification and biological evaluation of thiazole-based inverse agonists of ROR gamma t.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
4KZ5
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 5 (N-{[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl}glycine) | | Descriptor: | Beta-lactamase, N-{[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl}glycine, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4OWY
 
 | | Crystal Structure of Ahp1 from Saccharomyces cerevisiae. Investigating the electron transfers. | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Peroxiredoxin type-2, ... | | Authors: | Schultz, L, Genu, V, Breyer, C.A, dos Santos, V.F, Guimaraes, B.G, Netto, L.E.S, de Oliveira, M.A. | | Deposit date: | 2014-02-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Ahp1 from Saccharomyces cerevisiae. Investigating the electron transfers.
To be published
|
|
5CN4
 
 | | Ultrafast dynamics in myoglobin: -0.1 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
4DVZ
 
 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA
Cell Host Microbe, 12, 2012
|
|
3HJ8
 
 | | Crystal structure determination of catechol 1,2-dioxygenase from rhodococcus opacus 1CP in complex with 4-chlorocatechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 4-CHLOROBENZENE-1,2-DIOL, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-05-21 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts
J.Struct.Biol., 170, 2010
|
|
4CYQ
 
 | | Leishmania major N-myristoyltransferase in complex with a hybrid inhibitor (compound 45). | | Descriptor: | GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, N-{2-chloro-5-[(3S,4R)-1-[(3R)-4-(4-chlorophenyl)-3-hydroxybutanoyl]-4-(hydroxymethyl)pyrrolidin-3-yl]phenyl}-2-(4-fluorophenyl)acetamide, ... | | Authors: | Hutton, J.A, Goncalves, V, Brannigan, J.A, Paape, D, Waugh, T, Roberts, S.M, Bell, A.S, Wilkinson, A.J, Smith, D.F, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2014-04-14 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Based Design of Potent and Selective Leishmania N- Myristoyltransferase Inhibitors.
J.Med.Chem., 57, 2014
|
|
4LON
 
 | |
1KX8
 
 | | Antennal Chemosensory Protein A6 from Mamestra brassicae, tetragonal form | | Descriptor: | CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray Structure and Ligand Binding Study of a Chemosensory Protein
J.Biol.Chem., 277, 2002
|
|
4PEM
 
 | | Crystal Structure of S1G mutant of Penicillin G Acylase from Kluyvera citrophila | | Descriptor: | CALCIUM ION, Penicillin G acylase Alpha, Penicillin G acylase Beta | | Authors: | Ramasamy, S, Varshney, N.K, Brannigan, J.A, Wilkinson, A.J, Suresh, C.G. | | Deposit date: | 2014-04-24 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of S1G mutant of Penicillin G Acylase from Kluyvera citrophilla
To be Published
|
|
