2LO0
 
 | |
4C7I
 
 | | Leismania major N-myristoyltransferase in complex with a peptidomimetic (-OH) molecule | | Descriptor: | DIMETHYL SULFOXIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Olaleye, T.O, Brannigan, J.A, Goncalves, V, Roberts, S.M, Leatherbarrow, R.J, Wilkinson, A.J, Tate, E.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Peptidomimetic Inhibitors of N-Myristoyltransferase from Human Malaria and Leishmaniasis Parasites.
Org.Biomol.Chem., 12, 2014
|
|
4Z9O
 
 | | Crystal Structure of human GGT1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Terzyan, S.S, Hanigan, M.H. | | Deposit date: | 2015-04-10 | | Release date: | 2015-06-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human gamma-Glutamyl Transpeptidase 1: STRUCTURES OF THE FREE ENZYME, INHIBITOR-BOUND TETRAHEDRAL TRANSITION STATES, AND GLUTAMATE-BOUND ENZYME REVEAL NOVEL MOVEMENT WITHIN THE ACTIVE SITE DURING CATALYSIS.
J.Biol.Chem., 290, 2015
|
|
4CB7
 
 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound cap analogue (compound 8e) | | Descriptor: | 5-[2-(2-azanyl-7-methyl-6-oxidanylidene-1H-purin-9-ium-9-yl)ethanoyl]-2-oxidanyl-benzoic acid, GLYCEROL, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
4CB5
 
 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound cap analogue (compound 8f) | | Descriptor: | 9-N-(3-CARBOXY-4-HYDROXYPHENYL)KETOMETHYL-7-N-METHYLGUANINE, CHLORIDE ION, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
4CB6
 
 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound cap analogue (compound 11) | | Descriptor: | 2,9-N,N-DI(4-CARBOXYBUTYL)-7-N-METHYLGUANINE, IODIDE ION, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
4CB4
 
 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
6A69
 
 | | Cryo-EM structure of a P-type ATPase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroplastin, Plasma membrane calcium-transporting ATPase 1 | | Authors: | Gong, D.S, Chi, X.M, Ren, K, Huang, G.X.Y, Zhou, G.W, Yan, N, Lei, J.L, Zhou, Q. | | Deposit date: | 2018-06-27 | | Release date: | 2018-09-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Structure of the human plasma membrane Ca2+-ATPase 1 in complex with its obligatory subunit neuroplastin.
Nat Commun, 9, 2018
|
|
4KMI
 
 | | Crystal structure of 4-O-beta-D-mannosyl-D-glucose phosphorylase MGP complexed with PO4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-O-beta-D-mannosyl-D-glucose phosphorylase, PHOSPHATE ION | | Authors: | Nakae, S, Ito, S, Higa, M, Senoura, T, Wasaki, J, Hijikata, A, Shionyu, M, Ito, S, Shirai, T. | | Deposit date: | 2013-05-08 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Novel Enzyme in Mannan Biodegradation Process 4-O-beta-d-Mannosyl-d-Glucose Phosphorylase MGP
J.Mol.Biol., 425, 2013
|
|
1IDP
 
 | |
1TD5
 
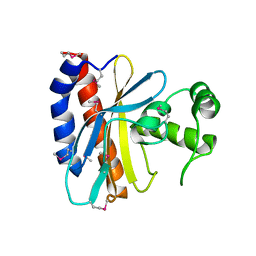 | | Crystal Structure of the Ligand Binding Domain of E. coli IclR. | | Descriptor: | Acetate operon repressor | | Authors: | Walker, J.R, Evdokimova, L, Zhang, R.-G, Bochkarev, A, Joachimiak, A, Arrowsmith, C, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-21 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analyses of the Ligand Binding Sites of the IclR family of transcriptional regulators
To be Published
|
|
3QMM
 
 | |
1IK8
 
 | | NMR structure of Alpha-Bungarotoxin | | Descriptor: | LONG NEUROTOXIN 1 | | Authors: | Niccolai, N, Ciutti, A, Spiga, O. | | Deposit date: | 2001-05-03 | | Release date: | 2001-05-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
4P1M
 
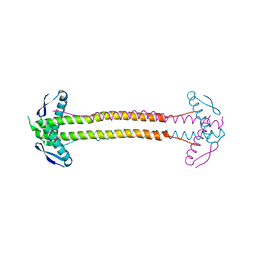 | | The structure of Escherichia coli ZapA | | Descriptor: | CHLORIDE ION, Cell division protein ZapA | | Authors: | Kimber, M.S, Roach, E.J, Khursigara, C.M. | | Deposit date: | 2014-02-26 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure and Site-directed Mutational Analysis Reveals Key Residues Involved in Escherichia coli ZapA Function.
J.Biol.Chem., 289, 2014
|
|
4KPW
 
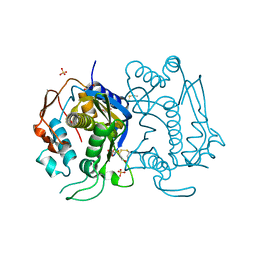 | |
6AHT
 
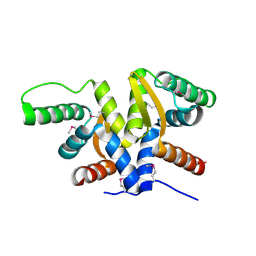 | | Plasmid partitioning protein TubR from Bacillus cereus | | Descriptor: | Conserved hypothetical plasmid protein | | Authors: | Hayashi, I. | | Deposit date: | 2018-08-20 | | Release date: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cooperative DNA Binding of the Plasmid Partitioning Protein TubR from the Bacillus cereus pXO1 Plasmid.
J.Mol.Biol., 430, 2018
|
|
3HHX
 
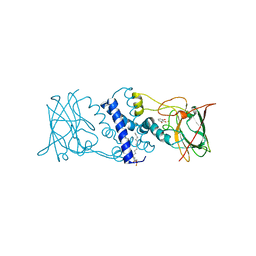 | | Crystal structure determination of Catechol 1,2-Dioxygenase from Rhodococcus opacus 1CP in complex with pyrogallol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BENZENE-1,2,3-TRIOL, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Briganti, F. | | Deposit date: | 2009-05-18 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
1IKC
 
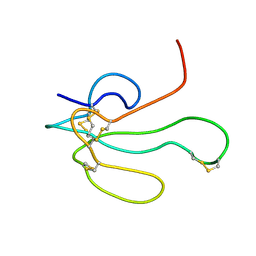 | | NMR Structure of alpha-Bungarotoxin | | Descriptor: | long neurotoxin 1 | | Authors: | Niccolai, N, Spiga, O, Ciutti, A. | | Deposit date: | 2001-05-03 | | Release date: | 2001-05-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
5ZYX
 
 | | Solution NMR structure of K30 peptide in 10 mM dioctanoyl phosphatidylglycerol (D8PG) | | Descriptor: | ARG-TRP-LYS-ARG-HIS-ILE-SER-GLU-GLN-LEU-ARG-ARG-ARG-ASP-ARG-LEU-GLN-ARG-GLN-ALA | | Authors: | Bhunia, A, Mohid, A, Stella, L, Calligari, P. | | Deposit date: | 2018-05-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design, Synthesis, Antibacterial Potential, and Structural Characterization of N-Acylated Derivatives of the Human Autophagy 16 Polypeptide.
Bioconjug.Chem., 30, 2019
|
|
4PU2
 
 | | Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine L-(R)-LeuP | | Descriptor: | Aminopeptidase N, GLYCEROL, LEUCINE PHOSPHONIC ACID, ... | | Authors: | Nocek, B, Vassiliou, S, Berlicki, L, Mulligan, R, Mucha, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-11 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine
To be Published
|
|
4PEL
 
 | | S1C mutant of Penicillin G acylase from Kluyvera citrophila | | Descriptor: | CALCIUM ION, Penicillin G acylase subunit alpha, Penicillin G acylase subunit beta | | Authors: | Ramasamy, S, Chand, D, Varshney, N.K, Brannigan, J.A, Wilkinson, A.J, Suresh, C.G. | | Deposit date: | 2014-04-24 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Penicillin G acylase
To Be Published
|
|
4KZ3
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 44 (5-chloro-3-sulfamoylthiophene-2-carboxylic acid) | | Descriptor: | 5-chloro-3-sulfamoylthiophene-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
3HHY
 
 | | Crystal structure determination of Catechol 1,2-Dioxygenase from Rhodococcus opacus 1CP in complex with catechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, CATECHOL, CHLORIDE ION, ... | | Authors: | Matera, I, Ferraroni, M, Kolomytseva, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-05-18 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
4PVB
 
 | | Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine (D-(S)-LeuP) | | Descriptor: | Aminopeptidase N, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Nocek, B, Vassiliou, S, Berlicki, L, Mulligan, R, Mucha, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-16 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine (D-(S)-LeuP)
To be Published
|
|
3HGI
 
 | | Crystal structure of Catechol 1,2-Dioxygenase from the gram-positive Rhodococcus opacus 1CP | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BENZOIC ACID, CARBONATE ION, ... | | Authors: | Ferraroni, M, Matera, I, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-05-14 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
