4TYV
 
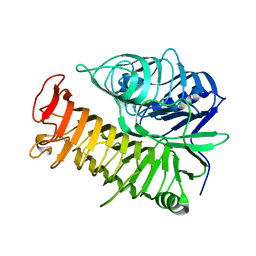 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
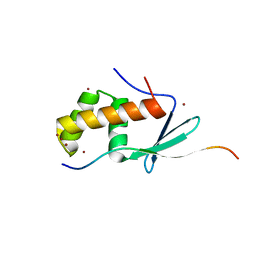
6JX3
 
 | |
6QTU
 
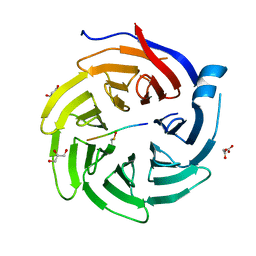 | |
6JYZ
 
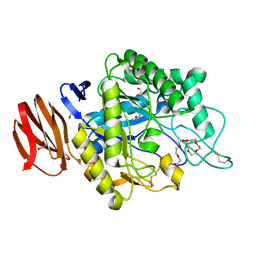 | | Crystal structure of endogalactoceramidase | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ISOPROPYL ALCOHOL, ... | | Authors: | Liuqing, C, Yan, F. | | Deposit date: | 2019-04-29 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of an endogalactosylceramidase from Rhodococcus hoagii 103S reveals the molecular basis of its substrate specificity.
J.Struct.Biol., 208, 2019
|
|
4UVV
 
 | | Crystal structure of human tankyrase 2 in complex with 3-(4- chlorophenyl)-5-methyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 3-(4-chlorophenyl)-5-methylisoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
6K3G
 
 | | Crystal structure of 10-Hydroxygeraniol Dehydrogenase from Cantharanthus roseus in complex with NADP+ | | Descriptor: | 10-hydroxygeraniol oxidoreductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Sandholu, A.S, Sharmila, P.M, Thulasiram, H.V, Kulkarni, K.A. | | Deposit date: | 2019-05-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural studies on 10-hydroxygeraniol dehydrogenase: A novel linear substrate-specific dehydrogenase from Catharanthus roseus.
Proteins, 88, 2020
|
|
4TUV
 
 | |
4U72
 
 | | Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase (A260G mutant) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-PHENYL-1H-IMIDAZOLE, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Sugimoto, H, Horitani, M, Kometani, E, Shiro, Y. | | Deposit date: | 2014-07-30 | | Release date: | 2015-09-02 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformation and Mobility of Active Site Loop is Critical for Substrate Binding and Inhibition in Human Indoleamine 2,3-Dioxygenase
to be published
|
|
3QJM
 
 | | Structural flexibility of Shank PDZ domain is important for its binding to different ligands | | Descriptor: | Beta-PIX, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Lee, J.H, Park, H, Park, S.J, Kim, H.J, Eom, S.H. | | Deposit date: | 2011-01-30 | | Release date: | 2011-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | The structural flexibility of the shank1 PDZ domain is important for its binding to different ligands
Biochem.Biophys.Res.Commun., 407, 2011
|
|
8EG0
 
 | |
4TVO
 
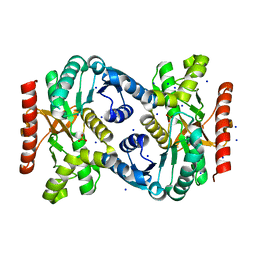 | |
6JWF
 
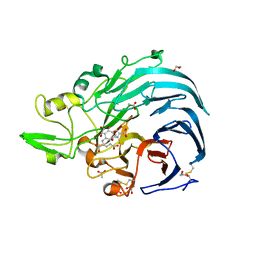 | | Holo form of Pyranose Dehydrogenase PQQ domain from Coprinopsis cinerea | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Takeda, K, Ishida, T, Yoshida, M, Samejima, M, Ohno, H, Igarashi, K, Nakamura, N. | | Deposit date: | 2019-04-20 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of the Catalytic and CytochromebDomains in a Eukaryotic Pyrroloquinoline Quinone-Dependent Dehydrogenase.
Appl.Environ.Microbiol., 85, 2019
|
|
4TZ7
 
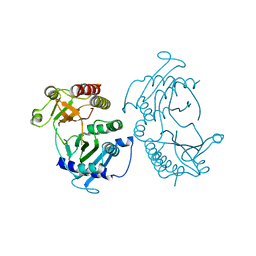 | | Crystal structure of type I phosphatidylinositol 4-phosphate 5-kinase alpha from Zebrafish | | Descriptor: | Phosphatidylinositol-4-phosphate 5-kinase, type I, alpha | | Authors: | Hu, J, Qin, Y, Wang, J, Li, L, Wu, D, Ha, Y. | | Deposit date: | 2014-07-09 | | Release date: | 2015-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Resolution of structure of PIP5K1A reveals molecular mechanism for its regulation by dimerization and dishevelled.
Nat Commun, 6, 2015
|
|
8DYB
 
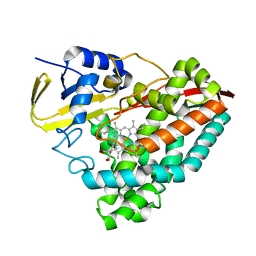 | |
6K9O
 
 | | Crystal Structure Analysis of Protein | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
8E1W
 
 | | Neutron crystal structure of Panus similis AA9A at room temperature | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Meilleur, F, Tandrup, T, Lo Leggio, L. | | Deposit date: | 2022-08-11 | | Release date: | 2023-01-11 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2.1 Å), X-RAY DIFFRACTION | | Cite: | Joint X-ray/neutron structure of Lentinus similis AA9_A at room temperature.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
6JZ7
 
 | |
4U9U
 
 | |
8E73
 
 | | Vigna radiata supercomplex I+III2 (full bridge) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Maldonado, M, Letts, J.A. | | Deposit date: | 2022-08-23 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Plant-specific features of respiratory supercomplex I + III 2 from Vigna radiata.
Nat.Plants, 9, 2023
|
|
4U2S
 
 | |
6QXU
 
 | | Human TNKS1 in complex with 6,8-Difluoro-2-[4-(1-hydroxy-1-methyl-ethyl)-phenyl]-3H-quinazolin-4-one | | Descriptor: | 6,8-bis(fluoranyl)-2-[4-(2-oxidanylpropan-2-yl)phenyl]-3~{H}-quinazolin-4-one, BETA-MERCAPTOETHANOL, Poly [ADP-ribose] polymerase tankyrase-1, ... | | Authors: | Musil, D, Lehmann, D, Buchstaller, H.-P. | | Deposit date: | 2019-03-08 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery and Optimization of 2-Arylquinazolin-4-ones into a Potent and Selective Tankyrase Inhibitor Modulating Wnt Pathway Activity.
J.Med.Chem., 62, 2019
|
|
4U3W
 
 | |
6KE3
 
 | |
6AAG
 
 | |
4WJQ
 
 | | Crystal Structure of SUMO1 in complex with Daxx | | Descriptor: | Daxx, Small ubiquitin-related modifier 1 | | Authors: | Cappadocia, L, Mascle, X.H, Bourdeau, V, Tremblay-Belzile, S, Chaker-Margot, M, Lussier-Price, M, Wada, J, Sakaguchi, K, Aubry, M, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Structure, 23, 2015
|
|
