8P2E
 
 | | Homotypic interacting B1 fab bound to Chondroitin Sulfate A | | Descriptor: | B1 fab heavy, B1 fab light | | Authors: | Raghavan, S.S.R, Dagil, R, Wang, K.T, Salanti, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Tumor-agnostic cancer therapy using antibodies targeting oncofetal chondroitin sulfate.
Nat Commun, 15, 2024
|
|
1ZTP
 
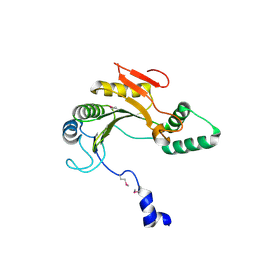 | | X-ray structure of gene product from homo sapiens Hs.433573 | | Descriptor: | Basophilic leukemia expressed protein BLES03 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-05-27 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure at 2.5 A resolution of human basophilic leukemia-expressed protein BLES03.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1VDF
 
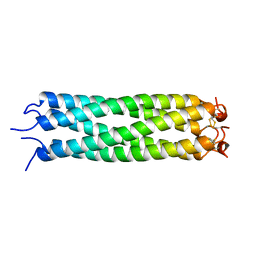 | |
8P29
 
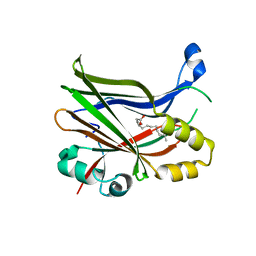 | | TEAD2 in complex with an inhibitor | | Descriptor: | 5-methyl-2-[(3-phenylmethoxyphenyl)amino]benzoic acid, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Guichou, J.F, Gelin, M, Allemand, F. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
1QOS
 
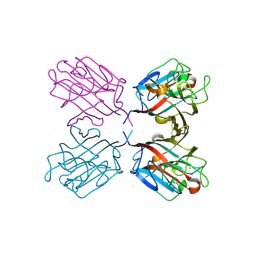 | | lectin UEA-II complexed with chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Loris, R, De Greve, H, Dao-Thi, M.-H, Messens, J, Imberty, A, Wyns, L. | | Deposit date: | 1999-11-16 | | Release date: | 2000-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by Lectin II from Ulex Europaeus, a Protein with a Promiscuous Carbohydrate Binding Site
J.Mol.Biol., 301, 2000
|
|
8TH7
 
 | |
8TH6
 
 | | Crystal Structure of the G3BP1 NTF2-like domain bound to USP10 peptide | | Descriptor: | 1,2-ETHANEDIOL, Ras GTPase-activating protein-binding protein 1, Ubiquitin carboxyl-terminal hydrolase 10 | | Authors: | Hughes, M.P, Taylor, J.P, Yang, Z. | | Deposit date: | 2023-07-14 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Interaction between host G3BP and viral nucleocapsid protein regulates SARS-CoV-2 replication and pathogenicity.
Cell Rep, 43, 2024
|
|
8TH5
 
 | |
8TNI
 
 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing bi-specific antibody CAP256L-R27 targeting the CD4-binding site and the V2-apex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 BG505 DS-SOSIP gp120, ... | | Authors: | Zhou, T, Morano, N.C, Roark, R.S, Kwong, P.D, Xu, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Ultrapotent Broadly Neutralizing Human-llama Bispecific Antibodies against HIV-1.
Adv Sci, 11, 2024
|
|
8TNH
 
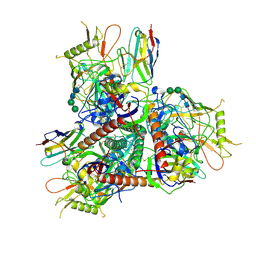 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing llama nanobody G36 targeting the CD4-binding site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD4-binding site nanobody G36, ... | | Authors: | Zhou, T, Kwong, P.D, Xu, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Ultrapotent Broadly Neutralizing Human-llama Bispecific Antibodies against HIV-1.
Adv Sci, 11, 2024
|
|
8TNG
 
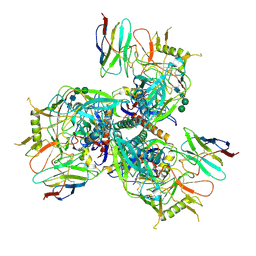 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing llama nanobody R27 targeting the CD4-binding site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD4-binding site targeting nanobody R27, ... | | Authors: | Zhou, T, Kwong, P.D, Xu, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Ultrapotent Broadly Neutralizing Human-llama Bispecific Antibodies against HIV-1.
Adv Sci, 11, 2024
|
|
2IS4
 
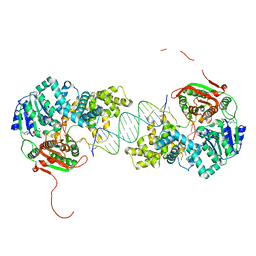 | | Crystal structure of UvrD-DNA-ADPNP ternary complex | | Descriptor: | 25-MER, DNA helicase II, MAGNESIUM ION, ... | | Authors: | Yang, W, Lee, J.Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke.
Cell(Cambridge,Mass.), 127, 2006
|
|
1JCR
 
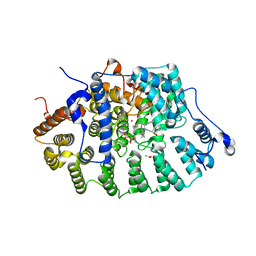 | | CRYSTAL STRUCTURE OF RAT PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH THE NON-SUBSTRATE TETRAPEPTIDE INHIBITOR CVFM AND FARNESYL DIPHOSPHATE SUBSTRATE | | Descriptor: | ACETIC ACID, FARNESYL DIPHOSPHATE, PROTEIN FARNESYLTRANSFERASE, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 2001-06-11 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human protein farnesyltransferase reveals the basis for inhibition by CaaX tetrapeptides and their mimetics.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7QM2
 
 | |
1JIE
 
 | | Crystal structure of bleomycin-binding protein from bleomycin-producing Streptomyces verticillus complexed with metal-free bleomycin | | Descriptor: | BLEOMYCIN A2, bleomycin-binding protein | | Authors: | Sugiyama, M, Kumagai, T, Hayashida, M, Maruyama, M, Matoba, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2002-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.6-A crystal structure of the copper(II)-bound bleomycin complexed with the bleomycin-binding protein from bleomycin-producing Streptomyces verticillus.
J.Biol.Chem., 277, 2002
|
|
3P0I
 
 | |
1JCQ
 
 | | CRYSTAL STRUCTURE OF HUMAN PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH FARNESYL DIPHOSPHATE AND THE PEPTIDOMIMETIC INHIBITOR L-739,750 | | Descriptor: | 2(S)-{2(S)-[2(R)-AMINO-3-MERCAPTO]PROPYLAMINO-3(S)-METHYL}PENTYLOXY-3-PHENYLPROPIONYLMETHIONINE SULFONE, ACETIC ACID, FARNESYL DIPHOSPHATE, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 2001-06-11 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of human protein farnesyltransferase reveals the basis for inhibition by CaaX tetrapeptides and their mimetics.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7R0N
 
 | | KRasG12C in complex with GDP and compound 2 | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-02-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C.
Cancer Discov, 12, 2022
|
|
7R0M
 
 | | KRasG12C in complex with GDP and JDQ443 | | Descriptor: | 1-[6-[4-(5-chloranyl-6-methyl-1~{H}-indazol-4-yl)-5-methyl-3-(1-methylindazol-5-yl)pyrazol-1-yl]-2-azaspiro[3.3]heptan-2-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-02-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C.
Cancer Discov, 12, 2022
|
|
7R0Q
 
 | | KRasG12C in complex with GDP and compound 3 | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-02-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C.
Cancer Discov, 12, 2022
|
|
4JEJ
 
 | | GGGPS from Flavobacterium johnsoniae | | Descriptor: | Geranylgeranylglyceryl phosphate synthase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Peterhoff, D, Beer, B, Rajendran, C, Kumpula, E.P, Kapetaniou, E, Guldan, H, Wierenga, R.K, Sterner, R, Babinger, P. | | Deposit date: | 2013-02-27 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A comprehensive analysis of the geranylgeranylglyceryl phosphate synthase enzyme family identifies novel members and reveals mechanisms of substrate specificity and quaternary structure organization.
Mol.Microbiol., 92, 2014
|
|
2IS6
 
 | | Crystal structure of UvrD-DNA-ADPMgF3 ternary complex | | Descriptor: | 5'-D(*CP*GP*AP*GP*CP*AP*CP*TP*GP*CP*AP*GP*TP*GP*CP*TP*CP*GP*TP*TP*GP*TP*TP*AP*T)-3', ADENOSINE-5'-DIPHOSPHATE, DNA helicase II, ... | | Authors: | Yang, W, Lee, J.Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke.
Cell(Cambridge,Mass.), 127, 2006
|
|
1RF8
 
 | | Solution structure of the yeast translation initiation factor eIF4E in complex with m7GDP and eIF4GI residues 393 to 490 | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic initiation factor 4F subunit p150, Eukaryotic translation initiation factor 4E, ... | | Authors: | Gross, J.D, Moerke, N.J, von der Haar, T, Lugovskoy, A.A, Sachs, A.B, McCarthy, J.E.G, Wagner, G. | | Deposit date: | 2003-11-07 | | Release date: | 2003-12-23 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E.
Cell(Cambridge,Mass.), 115, 2003
|
|
5N0H
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed meropenem - new refinement | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
8AAA
 
 | | Crystal structure of SARS-CoV-2 S RBD in complex with a stapled peptide | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, Spike protein S1, Stapled peptide | | Authors: | Brear, P, Chen, L, Gaynor, K, Harman, M, Dods, R, Hyvonen, M. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multivalent bicyclic peptides are an effective antiviral modality that can potently inhibit SARS-CoV-2.
Nat Commun, 14, 2023
|
|
