5JN9
 
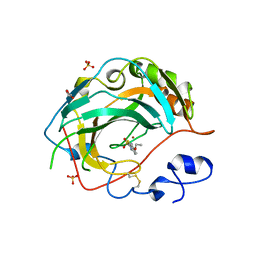 | | Crystal structure for the complex of human carbonic anhydrase IV and ethoxyzolamide | | Descriptor: | 6-ethoxy-1,3-benzothiazole-2-sulfonamide, ACETATE ION, Carbonic anhydrase 4, ... | | Authors: | Chen, Z, Waheed, A, Di Cera, E, Sly, W.S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
6F6S
 
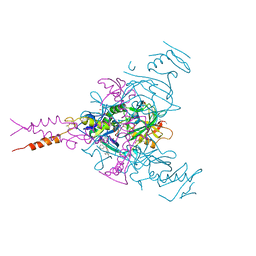 | | CRYSTAL STRUCTURE OF EBOLAVIRUS GLYCOPROTEIN IN COMPLEX WITH benztropine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Envelope glycoprotein, ... | | Authors: | Ren, J, Zhao, Y, Fry, E.E, Stuart, D.I. | | Deposit date: | 2017-12-06 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Target Identification and Mode of Action of Four Chemically Divergent Drugs against Ebolavirus Infection.
J. Med. Chem., 61, 2018
|
|
2WWB
 
 | | CRYO-EM STRUCTURE OF THE MAMMALIAN SEC61 COMPLEX BOUND TO THE ACTIVELY TRANSLATING WHEAT GERM 80S RIBOSOME | | Descriptor: | 25S RRNA, 5.8S RRNA, 60S RIBOSOMAL PROTEIN L17-A, ... | | Authors: | Becker, T, Mandon, E, Bhushan, S, Jarasch, A, Armache, J.P, Funes, S, Jossinet, F, Gumbart, J, Mielke, T, Berninghausen, O, Schulten, K, Westhof, E, Gilmore, R, Beckmann, R. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.48 Å) | | Cite: | Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome.
Science, 326, 2009
|
|
6OSA
 
 | | human Neurotensin Receptor 1 (hNTSR1) - Gi1 Protein Complex in non-canonical conformation (NC state) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Kato, H.E, Zhang, Y, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2019-05-01 | | Release date: | 2019-07-10 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational transitions of a neurotensin receptor 1-Gi1complex.
Nature, 572, 2019
|
|
6OS9
 
 | | human Neurotensin Receptor 1 (hNTSR1) - Gi1 Protein Complex in canonical conformation (C state) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Kato, H.E, Zhang, Y, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2019-05-01 | | Release date: | 2019-07-10 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational transitions of a neurotensin receptor 1-Gi1complex.
Nature, 572, 2019
|
|
7OFG
 
 | |
6FEC
 
 | | Human cap-dependent 48S pre-initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schaffitzel, C, Schaffitzel, C. | | Deposit date: | 2017-12-31 | | Release date: | 2018-03-14 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structure of a human cap-dependent 48S translation pre-initiation complex.
Nucleic Acids Res., 46, 2018
|
|
6VC4
 
 | | Peanut lectin complexed with S-beta-D-Thiogalactopyranosyl beta-D-glucopyranoside derivative (STGD) | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-6-{[(2S,3R,4S,5S,6S)-3,4,5-trihydroxy-6-({[(1-{[(2R,3S,4S,5R,6S)-3,4,5-trihydroxy-6-methoxytetrahydro-2H-pyran-2-yl]methyl}-1H-1,2,3-triazol-4-yl)methyl]sulfanyl}methyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-3,4,5-triol (non-preferred name), CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7PAZ
 
 | | REDUCED MUTANT P80I PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-21 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
5AII
 
 | | Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. CH55-sample-PEG complex | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Ferrandi, E, Sayer, C, Isupov, M.N, Annovazzi, C, Marchesi, C, Iacobone, G, Peng, X, Bonch-Osmolovskaya, E, Wohlgemuth, R, Littlechild, J.A, Montia, D. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries
FEBS J., 282, 2015
|
|
7QLH
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus amylovorus, domain I (aa 48-213) | | Descriptor: | PHOSPHATE ION, S-layer, SODIUM ION | | Authors: | Grininger, C, Sagmeister, T, Eder, E, Vejzovic, D, Pavkov-Keller, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QNM
 
 | | Crystallization and structural analyses of ZgHAD, a L-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans | | Descriptor: | (S)-2-haloacid dehalogenase, PHOSPHATE ION | | Authors: | Grigorian, E, Roret, T, Leblanc, C, Delage, L, Czjzek, M. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | X-ray structure and mechanism of ZgHAD, a l-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans.
Protein Sci., 32, 2023
|
|
3KS8
 
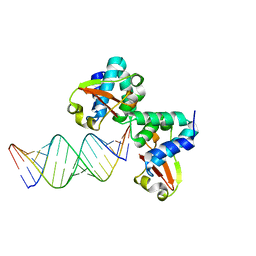 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain in complex with 18bp dsRNA | | Descriptor: | 5'-R(*AP*GP*AP*AP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*A)-3', 5'-R(*UP*CP*CP*UP*CP*CP*CP*UP*CP*CP*CP*UP*CP*CP*UP*UP*CP*U)-3', Polymerase cofactor VP35 | | Authors: | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
7QUX
 
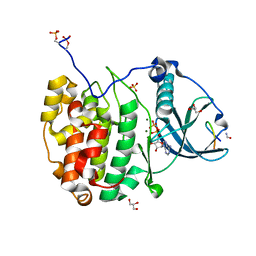 | | Crystal structure of P7C8 bound to CK2alpha | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMIC ACID, Casein kinase II subunit alpha, ... | | Authors: | Atkinson, E, Iegre, J, Brear, P, Baker, D, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2022-01-19 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Development of small cyclic peptides targeting the CK2 alpha / beta interface.
Chem.Commun.(Camb.), 58, 2022
|
|
6YI3
 
 | |
7TOJ
 
 | | Crystal structure of carbohydrate esterase CspAcXE, apoenzyme | | Descriptor: | CHLORIDE ION, SGNH/GDSL hydrolase family protein | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
6Z1M
 
 | | Structure of an Ancestral glycosidase (family 1) bound to heme | | Descriptor: | 1,2-ETHANEDIOL, Ancestral reconstructed glycosidase, GLYCEROL, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M, Gamiz-Arco, G, Gutierrez-Rus, L, Ibarra-Molero, B, Oshino, Y, Petrovic, D, Romero-Rivera, A, Seelig, B, Kamerlin, S.C.L, Gaucher, E.A. | | Deposit date: | 2020-05-14 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase.
Nat Commun, 12, 2021
|
|
3NUL
 
 | | Profilin I from Arabidopsis thaliana | | Descriptor: | GLYCEROL, PROFILIN I, SULFATE ION | | Authors: | Thorn, K, Christensen, H.E.M, Shigeta, R, Huddler, D, Chua, N.-H, Shalaby, L, Lindberg, U, Schutt, C.E. | | Deposit date: | 1996-11-27 | | Release date: | 1997-12-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a major allergen from plants.
Structure, 5, 1997
|
|
6VC2
 
 | | LRH-1 bound to SS-RJW100 and a fragment of the Tif2 Coactivator | | Descriptor: | (1S,3aS,6aS)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-ol, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2019-12-20 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Enantiomer-specific activities of an LRH-1 and SF-1 dual agonist.
Sci Rep, 10, 2020
|
|
6SU1
 
 | |
4WEM
 
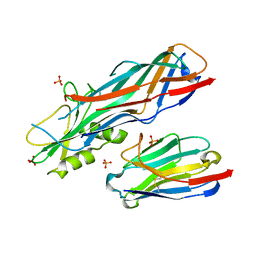 | | Co-complex structure of the F4 fimbrial adhesin FaeG variant ac with llama single domain antibody V1 | | Descriptor: | Anti-F4+ETEC bacteria VHH variable region, K88 fimbrial protein AC, PHOSPHATE ION | | Authors: | Moonens, K, Van den Broeck, I, Pardon, E, De Kerpel, M, Remaut, H, De Greve, H. | | Deposit date: | 2014-09-10 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight in the inhibition of adherence of F4 fimbriae producing enterotoxigenic Escherichia coli by llama single domain antibodies.
Vet. Res., 46, 2015
|
|
6MOA
 
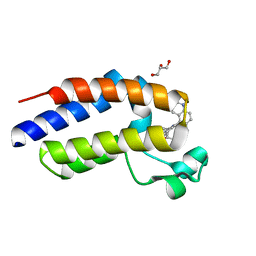 | | C-terminal bromodomain of human BRD2 in complex with 4-(2-cyclopropyl-7-(6-methylquinolin-5-yl)-1H-benzo[d]imidazol-5-yl)-3,5-dimethylisoxazole inhibitor | | Descriptor: | 4-(2-cyclopropyl-7-(6-methylquinolin-5-yl)-1H-benzo[d]imidazol-5-yl)-3,5-dimethylisoxazole, Bromodomain-containing protein 2, GLYCEROL | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6T14
 
 | | Native structure of mosquitocidal Cyt1A protoxin obtained by Serial Femtosecond Crystallography on in vivo grown crystals at pH 7 | | Descriptor: | SODIUM ION, Type-1Aa cytolytic delta-endotoxin | | Authors: | Tetreau, G, Banneville, A.S, Andreeva, E, Brewster, A.S, Hunter, M.S, Sierra, R.G, Young, I.D, Boutet, S, Coquelle, N, Cascio, D, Sawaya, M.R, Sauter, N.K, Colletier, J.P. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Serial femtosecond crystallography on in vivo-grown crystals drives elucidation of mosquitocidal Cyt1Aa bioactivation cascade.
Nat Commun, 11, 2020
|
|
6T1C
 
 | | Structure of the C7S mutant of mosquitocidal Cyt1A protoxin obtained by Serial Femtosecond Crystallography on in vivo grown crystals at pH 7 | | Descriptor: | SODIUM ION, Type-1Aa cytolytic delta-endotoxin | | Authors: | Tetreau, G, Banneville, A.S, Andreeva, E, Brewster, A.S, Hunter, M.S, Sierra, R.G, Young, I.D, Boutet, S, Coquelle, N, Cascio, D, Sawaya, M.R, Sauter, N.K, Colletier, J.P. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Serial femtosecond crystallography on in vivo-grown crystals drives elucidation of mosquitocidal Cyt1Aa bioactivation cascade.
Nat Commun, 11, 2020
|
|
6YWD
 
 | | De novo designed protein 4H_01 in complex with Mota antibody | | Descriptor: | Antibody Mota, Heavy Chain, Light Chain, ... | | Authors: | Yang, C, Sesterhenn, F, Pojer, F, Correia, B.E. | | Deposit date: | 2020-04-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Bottom-up de novo design of functional proteins with complex structural features.
Nat.Chem.Biol., 17, 2021
|
|
