6UF8
 
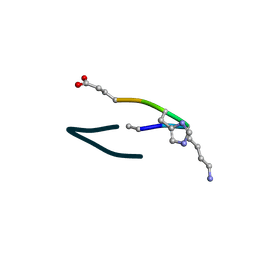 | | S2 symmetric peptide design number 6, London Bridge | | Descriptor: | S2-6, London Bridge | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
8E98
 
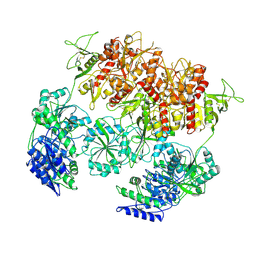 | | D-cycloserine and glutamate bound Human GluN1a-GluN2C NMDA receptor in nanodisc - intact conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E93
 
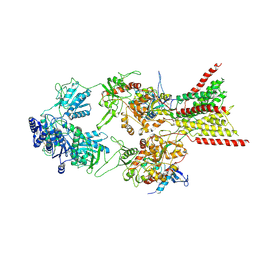 | | D-cycloserine and glutamate bound Human GluN1a-GluN2C NMDA receptor in splayed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E92
 
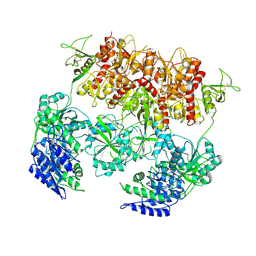 | | D-cycloserine and glutamate bound Human GluN1a-GluN2C NMDA receptor in intact conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
9DW6
 
 | |
9BJO
 
 | | Cryo-EM of Azo-ffsy fiber | | Descriptor: | D-peptide ffsy | | Authors: | Zia, A, Guo, J, Xu, B, Wang, F. | | Deposit date: | 2024-04-25 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cell-Free Nonequilibrium Assembly for Hierarchical Protein/Peptide Nanopillars.
J.Am.Chem.Soc., 146, 2024
|
|
9BJN
 
 | | Cryo-EM of Azo-ffspy fiber | | Descriptor: | D-peptide ffspy | | Authors: | Zia, A, Guo, J, Xu, B, Wang, F. | | Deposit date: | 2024-04-25 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cell-Free Nonequilibrium Assembly for Hierarchical Protein/Peptide Nanopillars.
J.Am.Chem.Soc., 146, 2024
|
|
6DZA
 
 | |
8Q7J
 
 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability | | Descriptor: | CYCLOSPORIN A | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
4XA8
 
 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2 | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding | | Authors: | Handing, K.B, Gasiorowska, O.A, Shabalin, I.G, Cymborowski, M.T, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-12-12 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Xanthobacter autotrophicus Py2.
to be published
|
|
1U6P
 
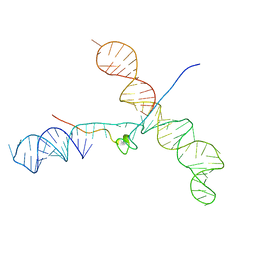 | |
6QYT
 
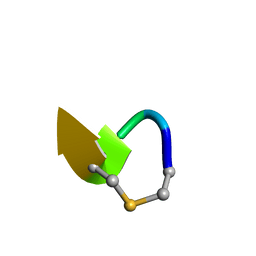 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring A truncated analogue | | Descriptor: | DAL-LEU-SER-LEU-CYS-ALA | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QYW
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - nisin ring A | | Descriptor: | ILE-DBU-DAL-ILE-DHA-LEU-CYS-ALA | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2019-10-02 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QYS
 
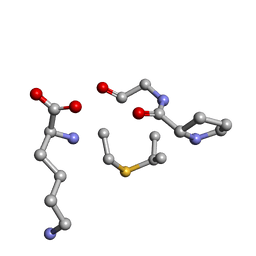 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Nisin Ring B | | Descriptor: | DBB-PRO-GLY-CYS-LYS | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
8EFQ
 
 | | DAMGO-bound mu-opioid receptor-Gi complex | | Descriptor: | DAMGO, ETHANOLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8CK0
 
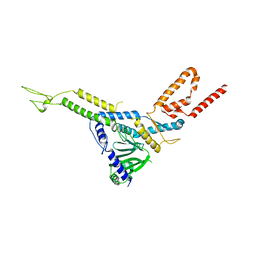 | |
6ZR9
 
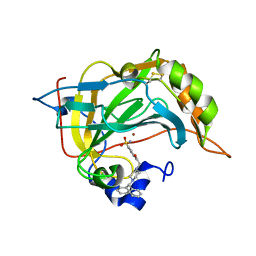 | |
6D2U
 
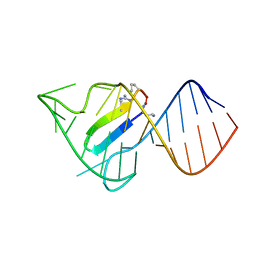 | |
8A3L
 
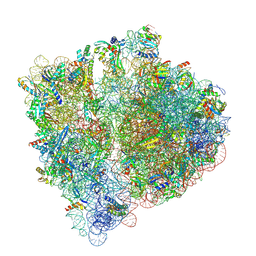 | | Structural insights into the binding of bS1 to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | D'Urso, G, Chat, S, Gillet, R, Giudice, E. | | Deposit date: | 2022-06-08 | | Release date: | 2023-05-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural insights into the binding of bS1 to the ribosome.
Nucleic Acids Res., 51, 2023
|
|
6WGN
 
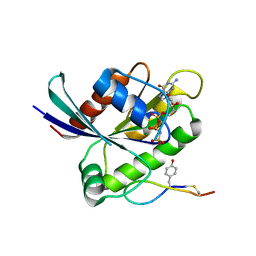 | | Crystal structure of K-Ras(G12D) GppNHp bound to cyclic peptide ligand KD2 | | Descriptor: | Cyclic Peptide KD2, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Zhang, Z, Gao, R, Hu, Q, Peacock, H, Peacock, D.M, Shokat, K.M, Suga, H. | | Deposit date: | 2020-04-06 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | GTP-State-Selective Cyclic Peptide Ligands of K-Ras(G12D) Block Its Interaction with Raf.
Acs Cent.Sci., 6, 2020
|
|
6DZ9
 
 | |
6QYV
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring A (Ser2, Ala5, Ala8) analogue | | Descriptor: | PHE-SER-DAL-LEU-ALA-LEU-CYS-ALA | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QTF
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring B, major conformer | | Descriptor: | DCY-LEU-GLY-ALA-THR | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-02-25 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QYU
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring A | | Descriptor: | PHE-DHA-DAL-LEU-DHA-LEU-CYS-ALA | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QYR
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring B, minor conformer | | Descriptor: | DAL-LEU-GLY-CYS-THR | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
