8W0V
 
 | |
8VDA
 
 | |
8W0W
 
 | |
8U24
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 9 | | Descriptor: | Green Fluorescent Protein Variant #9, ccGFP 9 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U78
 
 | | Structure of a N-Me-D-Gln4,Lys10-teixobactin analogue | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CHLORIDE ION, N-Methyl-D-Gln4,Lys10-teixobactin | | Authors: | Nowick, J.S, Yang, H, Kreutzer, A.G. | | Deposit date: | 2023-09-14 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Supramolecular Interactions of Teixobactin Analogues in the Crystal State.
J.Org.Chem., 89, 2024
|
|
8UN9
 
 | |
1EGS
 
 | |
9BRT
 
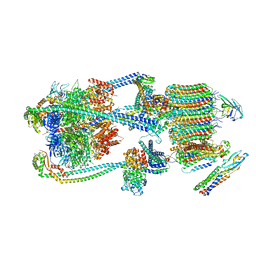 | | Intact V-ATPase State 1 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 2024
|
|
1E50
 
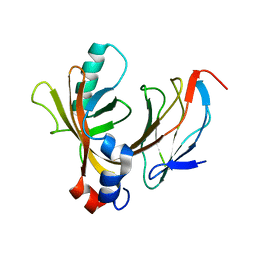 | | AML1/CBFbeta complex | | Descriptor: | CORE-BINDING FACTOR ALPHA SUBUNIT, CORE-BINDING FACTOR CBF-BETA | | Authors: | Warren, A.J, Bravo, J, Williams, R.L, Rabbits, T.H. | | Deposit date: | 2000-07-13 | | Release date: | 2001-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Heterodimeric Interaction between the Acute Leukaemia-Associated Transcription Factors Aml1 and Cbfbeta
Embo J., 19, 2000
|
|
8V96
 
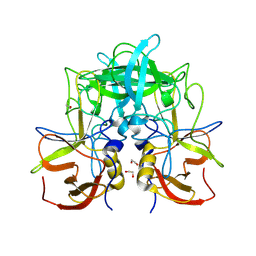 | | GII.14 M7 norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein VP1 | | Authors: | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 2024
|
|
9BRZ
 
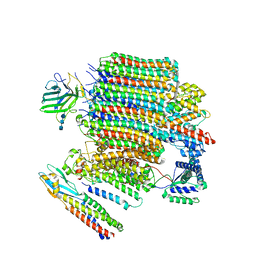 | | V0-only V-ATPase and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-12 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 2024
|
|
8XMS
 
 | |
8UTB
 
 | | Alpha7-nicotinic acetylcholine receptor bound to epibatidine and NS-1738 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
8V7W
 
 | |
8V8E
 
 | | Room-temperature X-ray structure of SARS-CoV-2 main protease catalytic domain (residues 1-199-6H) in complex with ensitrelvir (ESV) | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, peptide | | Authors: | Kovalevsky, A, Coates, L. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Visualizing the Active Site Oxyanion Loop Transition Upon Ensitrelvir Binding and Transient Dimerization of SARS-CoV-2 Main Protease.
J.Mol.Biol., 436, 2024
|
|
8W0X
 
 | |
8YPJ
 
 | |
8UT1
 
 | | Alpha7-nicotinic acetylcholine receptor bound to epibatidine | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Burke, S.M, Hibbs, R.E, Noviello, C.M. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
8VD9
 
 | |
8V98
 
 | | GII.24 Loreto 1972 norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Capsid protein VP1 | | Authors: | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 2024
|
|
8VDB
 
 | |
8V8C
 
 | | Alpha7-nicotinic acetylcholine receptor time resolved bound to epibatidine and PNU-120596 asymmetric state 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
8V95
 
 | | GII.8 Amsterdam norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein (Fragment) | | Authors: | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 2024
|
|
8TWS
 
 | | AvrB bound with UDP-rhamnose and RIN4 C-NOI motif | | Descriptor: | Avirulence protein B, RPM1-interacting protein 4, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{R},6~{S})-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Peng, W, Orth, K. | | Deposit date: | 2023-08-21 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Pseudomonas effector AvrB is a glycosyltransferase that rhamnosylates plant guardee protein RIN4.
Sci Adv, 10, 2024
|
|
8V7T
 
 | |
