3FRW
 
 | | Crystal structure of putative TrpR protein from Ruminococcus obeum | | Descriptor: | ACETATE ION, Putative Trp repressor protein | | Authors: | Osipiuk, J, Keigher, L, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-08 | | Release date: | 2009-01-20 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of putative TrpR protein from Ruminococcus obeum.
To be Published
|
|
4JFJ
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with compound (1S,6R)-10-(1,3-benzothiazol-6-ylsulfonyl)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-10-(1,3-benzothiazol-6-ylsulfonyl)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JG6
 
 | | RSK2 CTD bound to 2-cyano-3-(1H-indazol-5-yl)acrylamide | | Descriptor: | (2S)-2-cyano-3-(1H-indazol-5-yl)propanamide, Ribosomal protein S6 kinase alpha-3, SODIUM ION | | Authors: | Miller, R.M, Paavilainen, V.O, Krishnan, S, Serafimova, I.M, Taunton, J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-04-10 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Electrophilic fragment-based design of reversible covalent kinase inhibitors.
J.Am.Chem.Soc., 135, 2013
|
|
3OXF
 
 | | Human lysine methyltransferase Smyd3 in complex with AdoHcy (Form I) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SET and MYND domain-containing protein 3, ZINC ION | | Authors: | Xu, S, Wu, J, Sun, B, Zhong, C, Ding, J. | | Deposit date: | 2010-09-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural and biochemical studies of human lysine methyltransferase Smyd3 reveal the important functional roles of its post-SET and TPR domains and the regulation of its activity by DNA binding.
Nucleic Acids Res., 39, 2011
|
|
4NXM
 
 | | Crystal Structure of the 30S ribosomal subunit from a GidB (RsmG) mutant of Thermus thermophilus (HB8) | | Descriptor: | 16S rRNA, MAGNESIUM ION, ZINC ION, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2013-12-09 | | Release date: | 2014-05-21 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
4JGE
 
 | |
2NZ5
 
 | | Structure and Function Studies of Cytochrome P450 158A1 from Streptomyces coelicolor A3(2) | | Descriptor: | Cytochrome P450 CYP158A1, PROTOPORPHYRIN IX CONTAINING FE, naphthalene-1,2,4,5,7-pentol | | Authors: | Zhao, B, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2006-11-22 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Different binding modes of two flaviolin substrate molecules in cytochrome P450 158A1 (CYP158A1) compared to CYP158A2.
Biochemistry, 46, 2007
|
|
2NZT
 
 | | Crystal structure of human hexokinase II | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, Hexokinase-2, UNKNOWN ATOM OR ION, ... | | Authors: | Rabeh, W.M, Zhu, H, Nedyalkova, L, Tempel, W, Wasney, G, Landry, R, Vedadi, M, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-25 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The catalytic inactivation of the N-half of human hexokinase 2 and structural and biochemical characterization of its mitochondrial conformation.
Biosci.Rep., 38, 2018
|
|
4JH3
 
 | | Crystal Structure of FosB from Bacillus cereus with Zinc and Fosfomycin | | Descriptor: | FORMIC ACID, FOSFOMYCIN, GLYCEROL, ... | | Authors: | Thompson, M.K, Harp, J, Keithly, M.E, Jagessar, K, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-03-04 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Chemical Aspects of Resistance to the Antibiotic Fosfomycin Conferred by FosB from Bacillus cereus.
Biochemistry, 52, 2013
|
|
4O2I
 
 | | The crystal structure of non-LEE encoded type III effector C from Citrobacter rodentium | | Descriptor: | Non-LEE encoded type III effector C, ZINC ION | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of non-LEE encoded type III effector C from Citrobacter rodentium
To be Published
|
|
3FQC
 
 | | Staphylococcus aureus dihydrofolate reductase complexed with NADPH and 2,4-diamino-5-[3-(3,4,5-trimethoxyphenyl)pent-1-ynyl]-6-methylpyrimidine (UCP115A) | | Descriptor: | 6-methyl-5-[3-methyl-3-(3,4,5-trimethoxyphenyl)but-1-yn-1-yl]pyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethoprim-sensitive dihydrofolate reductase | | Authors: | Anderson, A.C, Frey, K.M, Liu, J, Lombardo, M.N. | | Deposit date: | 2009-01-07 | | Release date: | 2009-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of wild-type and mutant methicillin-resistant Staphylococcus aureus dihydrofolate reductase reveal an alternate conformation of NADPH that may be linked to trimethoprim resistance.
J.Mol.Biol., 387, 2009
|
|
4JHI
 
 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with N6-benzyladenine | | Descriptor: | MtN13 protein, N-BENZYL-9H-PURIN-6-AMINE, SODIUM ION | | Authors: | Ruszkowski, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3OP5
 
 | | Human vaccinia-related kinase 1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Serine/threonine-protein kinase VRK1, ... | | Authors: | Allerston, C.K, Uttarkar, S, Savitsky, P, Elkins, J.M, Filippakopoulos, P, Krojer, T, Rellos, P, Fedorov, O, Eswaran, J, Brenner, B, Keates, T, Das, S, King, O, Chalk, R, Berridge, G, von Delft, F, Gileadi, O, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of human Vaccinia-Related Kinases (VRK) bound to small-molecule inhibitors identifies different P-loop conformations.
Sci Rep, 7, 2017
|
|
4JHS
 
 | | Crystal structure of a C-terminal two domain fragment of human beta-2-glycoprotein 1 | | Descriptor: | Beta-2-glycoprotein 1, SULFATE ION | | Authors: | Bonanno, J.B, Toro, R, Gizzi, A, Chan, M.K, Garrett-Thomson, S.C, Patel, H, Lim, S, Matikainen, B, Celikgil, A, Garforth, S, Hillerich, B, Seidel, R, Rand, J.H, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a C-terminal two domain fragment of human beta-2-glycoprotein 1
To be Published
|
|
3FQN
 
 | |
4O3Z
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) mutant Tyr-95-Ala from Actinobacillus pleuropneumoniae H87 | | Descriptor: | ACETATE ION, GLYCEROL, Outer membrane protein; transferrin-binding protein | | Authors: | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | Deposit date: | 2013-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
4NX8
 
 | | Structure of a PTP-like phytase from Bdellovibrio bacteriovorus | | Descriptor: | GLYCEROL, MAGNESIUM ION, Protein-tyrosine phosphatase 2 | | Authors: | Gruninger, R.J, Lovering, A.L. | | Deposit date: | 2013-12-08 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structural and biochemical analysis of a unique phosphatase from Bdellovibrio bacteriovorus reveals its structural and functional relationship with the protein tyrosine phosphatase class of phytase.
Plos One, 9, 2014
|
|
4O4G
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in complex with 4-((4-(mesitylamino)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ527), a non-nucleoside inhibitor | | Descriptor: | 4-({4-[(2,4,6-trimethylphenyl)amino]-1,3,5-triazin-2-yl}amino)benzonitrile, HIV-1 reverse transcriptase, p51 subunit, ... | | Authors: | Mislak, A.C, Frey, K.M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | A mechanistic and structural investigation of modified derivatives of the diaryltriazine class of NNRTIs targeting HIV-1 reverse transcriptase.
Biochim.Biophys.Acta, 1840, 2014
|
|
4JC8
 
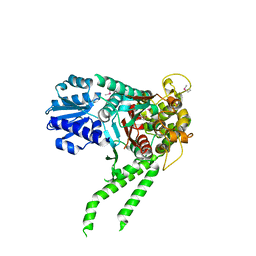 | |
4JI6
 
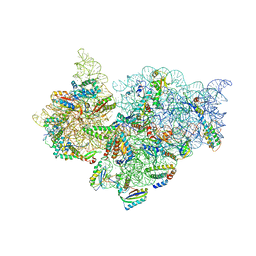 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, RIBOSOMAL PROTEIN S10, ... | | Authors: | Demirci, H, Wang, L, Murphy IV, F, Murphy, E, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2013-12-04 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
4NDH
 
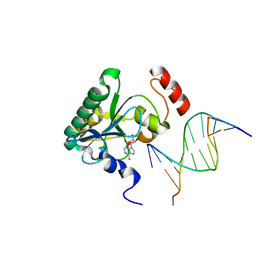 | | Human Aprataxin (Aptx) bound to DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(P*GP*TP*TP*CP*TP*AP*GP*AP*AP*C)-3', ADENOSINE MONOPHOSPHATE, Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4JEH
 
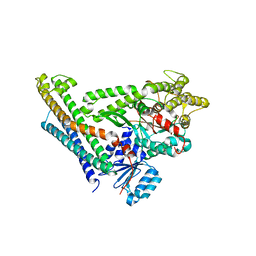 | | Crystal Structure of Munc18a and Syntaxin1 lacking N-peptide complex | | Descriptor: | Syntaxin-1A, Syntaxin-binding protein 1 | | Authors: | Colbert, K.N, Hattendorf, D.A, Weiss, T.M, Burkhardt, P, Fasshauer, D, Weis, W.I. | | Deposit date: | 2013-02-27 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Syntaxin1a variants lacking an N-peptide or bearing the LE mutation bind to Munc18a in a closed conformation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JF3
 
 | |
3FUT
 
 | | Apo-form of T. thermophilus 16S rRNA A1518 and A1519 methyltransferase (KsgA) in space group P21212 | | Descriptor: | Dimethyladenosine transferase | | Authors: | Demirci, H, Belardinelli, R, Seri, E, Gregory, S.T, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-01-14 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural rearrangements in the active site of the Thermus thermophilus 16S rRNA methyltransferase KsgA in a binary complex with 5'-methylthioadenosine.
J.Mol.Biol., 388, 2009
|
|
3OU9
 
 | |
