2Q54
 
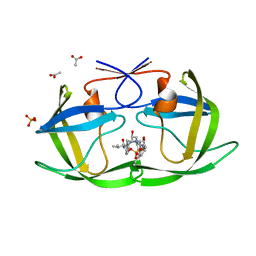 | | Crystal structure of KB73 bound to HIV-1 protease | | Descriptor: | ACETATE ION, N~2~-ACETYL-N-[(1S,3S,4S)-4-({[(5S)-3-(3-ACETYLPHENYL)-2-OXO-1,3-OXAZOLIDIN-5-YL]CARBONYL}AMINO)-1-BENZYL-3-HYDROXY-5-PHENYLPENTYL]-L-VALINAMIDE, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-05-31 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and Synthesis of HIV-1 Protease Inhibitors Incorporating Oxazolidinones as P2/P2' Ligands in Pseudosymmetric Dipeptide Isosteres.
J.Med.Chem., 50, 2007
|
|
2PQZ
 
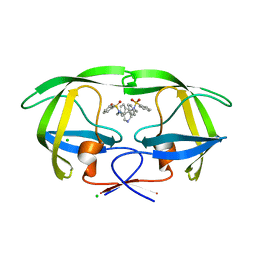 | | HIV-1 Protease in complex with a pyrrolidine-based inhibitor | | Descriptor: | CHLORIDE ION, N,N'-(3S,4S)-PYRROLIDINE-3,4-DIYLBIS(N-BENZYLBENZENESULFONAMIDE), Protease | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-05-03 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold.
J.Med.Chem., 51, 2008
|
|
2PVM
 
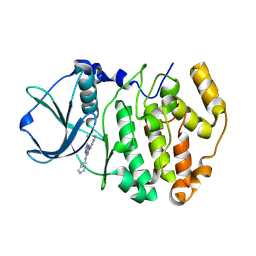 | | Structure-Based Design of Pyrazolo[1,5-a][1,3,5]triazine Derivatives as Potent Inhibitors of Protein Kinase CK2 | | Descriptor: | 4-(2-(1H-IMIDAZOL-4-YL)ETHYLAMINO)-2-(PHENYLAMINO)PYRAZOLO[1,5-A][1,3,5]TRIAZINE-8-CARBONITRILE, Casein kinase II subunit alpha | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Almassy, R, Lu, J, Averill, A, Yager, K.M, Chu, S. | | Deposit date: | 2007-05-09 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2PWA
 
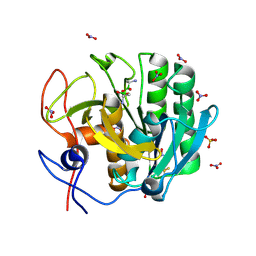 | | Crystal Structure of the complex of Proteinase K with Alanine Boronic acid at 0.83A resolution | | Descriptor: | ALANINE BORONIC ACID, CALCIUM ION, NITRATE ION, ... | | Authors: | Jain, R, Singh, N, Perbandt, M, Betzel, C, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-05-11 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | Crystal structure of the complex of Proteinase K with Alanine Boronic Acid at 0.83A Resolution
To be Published
|
|
2PWR
 
 | | HIV-1 protease in complex with a carbamoyl decorated pyrrolidine-based inhibitor | | Descriptor: | 4,4'-{(3S,4S)-PYRROLIDINE-3,4-DIYLBIS[(BENZYLIMINO)SULFONYL]}DIBENZAMIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Boettcher, B, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-05-12 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold.
J.Med.Chem., 51, 2008
|
|
2PWS
 
 | | Crystal structure of the complex formed between phospholipase A2 and 2-(4-isobutyl-phenyl)-propionic acid at 2.2 A resolution | | Descriptor: | IBUPROFEN, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Kumar, S, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-05-13 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the complex formed between phospholipase A2 and 2-(4-isobutyl-phenyl)-propionic acid at 2.2 resolution
To be Published
|
|
2PY9
 
 | |
2Q0A
 
 | | Structure and rearrangements in the carboxy-terminal region of SpIH channels | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Flynn, G.E, Black, K.D, Islas, L.D, Sankaran, B, Zagotta, W.N. | | Deposit date: | 2007-05-21 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and rearrangements in the carboxy-terminal region of SpIH channels.
Structure, 15, 2007
|
|
2Q0M
 
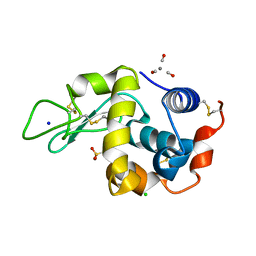 | | Tricarbonylmanganese(I)-lysozyme complex : a structurally characterized organometallic protein | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, Lysozyme C, ... | | Authors: | Razavet, M, Artero, V, Cavazza, C, Oudart, Y, Fontecilla-Camps, J.C, Fontecave, M. | | Deposit date: | 2007-05-22 | | Release date: | 2007-12-11 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tricarbonylmanganese(I)-lysozyme complex: a structurally characterized organometallic protein.
Chem.Commun.(Camb.), 2007
|
|
2Q15
 
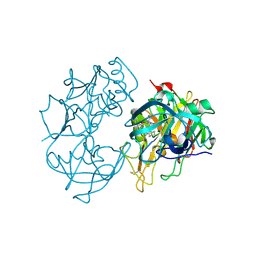 | | Structure of BACE complexed to compound 3a | | Descriptor: | (4S)-4-(2-AMINO-6-PHENOXYQUINAZOLIN-3(4H)-YL)-N,4-DICYCLOHEXYL-N-METHYLBUTANAMIDE, Beta-secretase 1 | | Authors: | Sharff, A.J. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-Amino-3,4-dihydroquinazolines as inhibitors of BACE-1 (beta-Site APP cleaving enzyme): Use of structure based design to convert a micromolar hit into a nanomolar lead.
J.Med.Chem., 50, 2007
|
|
2Q11
 
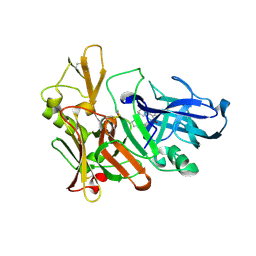 | | Structure of BACE complexed to compound 1 | | Descriptor: | 3-(2-AMINO-6-BENZOYLQUINAZOLIN-3(4H)-YL)-N-CYCLOHEXYL-N-METHYLPROPANAMIDE, Beta-secretase 1 | | Authors: | Sharff, A.J. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-Amino-3,4-dihydroquinazolines as inhibitors of BACE-1 (beta-Site APP cleaving enzyme): Use of structure based design to convert a micromolar hit into a nanomolar lead.
J.Med.Chem., 50, 2007
|
|
2Q5K
 
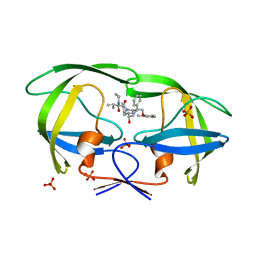 | | Crystal structure of lopinavir bound to wild type HIV-1 protease | | Descriptor: | N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-06-01 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design and Synthesis of HIV-1 Protease Inhibitors Incorporating Oxazolidinones as P2/P2' Ligands in Pseudosymmetric Dipeptide Isosteres.
J.Med.Chem., 50, 2007
|
|
5P5B
 
 | |
1VVD
 
 | |
5P5Q
 
 | |
5P63
 
 | |
1VVE
 
 | |
5P6F
 
 | |
2B60
 
 | | Structure of HIV-1 protease mutant bound to Ritonavir | | Descriptor: | GLYCEROL, Gag-Pol polyprotein, RITONAVIR | | Authors: | Clemente, J.C, Stow, L.R, Janka, L.K, Jeung, J.A, Desai, K.A, Govindasamy, L, Agbandje-McKenna, M, McKenna, R, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2005-09-29 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In vivo, kinetic, and structural analysis of the development of ritonavir resistance
To be Published
|
|
2YDK
 
 | | Discovery of Checkpoint Kinase Inhibitor AZD7762 by Structure Based Design and Optimization of Thiophene Carboxamide Ureas | | Descriptor: | 2-(CARBAMOYLAMINO)-5-PHENYL-N-[(3S)-PIPERIDIN-3-YL]THIOPHENE-3-CARBOXAMIDE, GLYCEROL, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Rowsell, S, Vallentine, A, White, A. | | Deposit date: | 2011-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(Piperidin-3-Yl)-3-Ureidothiophene-2-Carboxamide (Azd7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
3TKH
 
 | | Crystal structure of Chk1 in complex with inhibitor S01 | | Descriptor: | 1-(morpholin-4-yl)-2-[4-(2-{[5-(pyridin-3-yl)-1,3-thiazol-2-yl]amino}pyridin-4-yl)piperazin-1-yl]ethanone, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Ikuta, M. | | Deposit date: | 2011-08-26 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Pyridyl aminothiazoles as potent inhibitors of Chk1 with slow dissociation rates.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5P6U
 
 | |
1HUI
 
 | | INSULIN MUTANT (B1, B10, B16, B27)GLU, DES-B30, NMR, 25 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Olsen, H.B, Ludvigsen, S, Kaarsholm, N.C. | | Deposit date: | 1996-03-29 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an engineered insulin monomer at neutral pH.
Biochemistry, 35, 1996
|
|
5P79
 
 | |
1HUM
 
 | |
