3Q1N
 
 | |
2CCE
 
 | | Parallel Configuration of pLI E20S | | Descriptor: | General control protein GCN4 | | Authors: | Yadav, M.K, Leman, L.J, Price, D.J, Brooks 3rd, C.L, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2006-01-16 | | Release date: | 2006-04-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coiled coils at the edge of configurational heterogeneity. Structural analyses of parallel and antiparallel homotetrameric coiled coils reveal configurational sensitivity to a single solvent-exposed amino acid substitution.
Biochemistry, 45, 2006
|
|
2BGQ
 
 | | apo aldose reductase from barley | | Descriptor: | ALDOSE REDUCTASE, SULFATE ION | | Authors: | Olsen, J.G, Pedersen, L, Christensen, C.L, Olsen, O, Henriksen, A. | | Deposit date: | 2005-01-04 | | Release date: | 2006-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Barley Aldose Reductase: Structure, Cofactor Binding, and Substrate Recognition in the Aldo/Keto Reductase 4C Family.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
2BGS
 
 | | HOLO ALDOSE REDUCTASE FROM BARLEY | | Descriptor: | ALDOSE REDUCTASE, BICARBONATE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Olsen, J.G, Pedersen, L, Christensen, C.L, Olsen, O, Henriksen, A. | | Deposit date: | 2005-01-05 | | Release date: | 2006-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Barley Aldose Reductase: Structure, Cofactor Binding, and Substrate Recognition in the Aldo/Keto Reductase 4C Family.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
1ICT
 
 | | MONOCLINIC FORM OF HUMAN TRANSTHYRETIN COMPLEXED WITH THYROXINE (T4) | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, TRANSTHYRETIN | | Authors: | Wojtczak, A, Neumann, P, Cody, V. | | Deposit date: | 2001-04-02 | | Release date: | 2002-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a new polymorphic monoclinic form of human transthyretin at 3 A resolution reveals a mixed complex between unliganded and T4-bound tetramers of TTR.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3PF3
 
 | | Crystal structure of a mutant (C202A) of Triosephosphate isomerase from Giardia lamblia derivatized with MMTS | | Descriptor: | CALCIUM ION, GLYCEROL, SULFATE ION, ... | | Authors: | Enriquez-Flores, S, Rodriguez-Romero, A, Hernandez-Santoyo, A, Reyes-Vivas, H. | | Deposit date: | 2010-10-27 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Determining the molecular mechanism of inactivation by chemical modification of triosephosphate isomerase from the human parasite Giardia lamblia: A study for antiparasitic drug design.
Proteins, 79, 2011
|
|
3QJY
 
 | | Crystal structure of P-loop G234A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-31 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
1OT4
 
 | | Solution structure of Cu(II)-CopC from Pseudomonas syringae | | Descriptor: | COPPER (II) ION, Copper resistance protein C | | Authors: | Arnesano, F, Banci, L, Bertini, I, Felli, I.C, Luchinat, C, Thompsett, A.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-21 | | Release date: | 2003-07-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Strategy for the NMR Characterization of Type II Copper(II) Proteins:
the Case of the Copper Trafficking Protein CopC from Pseudomonas Syringae.
J.Am.Chem.Soc., 125, 2003
|
|
3QIA
 
 | | Crystal structure of P-loop G237A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-26 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3Q9T
 
 | | Crystal structure analysis of formate oxidase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Doubayashi, D, Ootake, T, Maeda, Y, Oki, M, Tokunaga, Y, Sakurai, A, Nagaosa, Y, Mikami, B, Uchida, H. | | Deposit date: | 2011-01-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Formate oxidase, an enzyme of the glucose-methanol-choline oxidoreductase family, has a His-Arg pair and 8-formyl-FAD at the catalytic site.
Biosci.Biotechnol.Biochem., 75, 2011
|
|
2CPQ
 
 | | Solution structure of the N-terminal KH domain of human FXR1 | | Descriptor: | Fragile X mental retardation syndrome related protein 1, isoform b' | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal KH domain of human FXR1
To be Published
|
|
2H8B
 
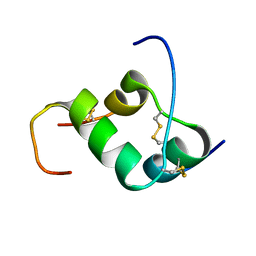 | | Solution structure of INSL3 | | Descriptor: | Insulin-like 3 | | Authors: | Rosengren, K.J, Craik, D.J, Daly, N.L. | | Deposit date: | 2006-06-07 | | Release date: | 2006-08-01 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Characterization of the LGR8 Receptor Binding Surface of Insulin-like Peptide 3
J.Biol.Chem., 281, 2006
|
|
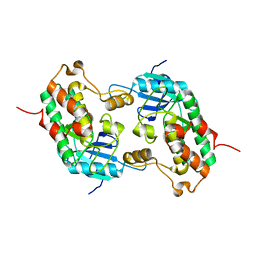
1ZWS
 
 | |
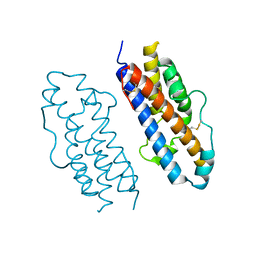
1HUW
 
 | |
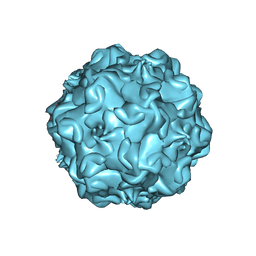
2CAS
 
 | |
2C7W
 
 | | Crystal Structure of human vascular endothelial growth factor-B: Identification of amino acids important for angiogeninc activity | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, VASCULAR ENDOTHELIAL GROWTH FACTOR B PRECURSOR | | Authors: | Iyer, S, Scotney, P.D, Nash, A.D, Acharya, K.R. | | Deposit date: | 2005-11-29 | | Release date: | 2007-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of Human Vascular Endothelial Growth Factor-B: Identification of Amino Acids Important for Receptor Binding
J.Mol.Biol., 359, 2006
|
|
3P2K
 
 | | Structure of an antibiotic related Methyltransferase | | Descriptor: | 16S rRNA methylase, S-ADENOSYLMETHIONINE | | Authors: | Sivaraman, J, Husain, N. | | Deposit date: | 2010-10-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the methylation of A1408 in 16S rRNA by a panaminoglycoside resistance methyltransferase NpmA from a clinical isolate and analysis of the NpmA interactions with the 30S ribosomal subunit.
Nucleic Acids Res., 39, 2011
|
|
1CEK
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE MEMBRANE-EMBEDDED M2 CHANNEL-LINING SEGMENT FROM THE NICOTINIC ACETYLCHOLINE RECEPTOR BY SOLID-STATE NMR SPECTROSCOPY | | Descriptor: | PROTEIN (ACETYLCHOLINE RECEPTOR M2) | | Authors: | Marassi, F.M, Gesell, J.J, Kim, Y, Valente, A.P, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 1999-03-09 | | Release date: | 1999-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLID-STATE NMR | | Cite: | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
2CCF
 
 | | Antiparallel Configuration of pLI E20S | | Descriptor: | General control protein GCN4 | | Authors: | Yadav, M.K, Leman, L.J, Price, D.J, Brooks 3rd, C.L, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2006-01-16 | | Release date: | 2006-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Coiled coils at the edge of configurational heterogeneity. Structural analyses of parallel and antiparallel homotetrameric coiled coils reveal configurational sensitivity to a single solvent-exposed amino acid substitution.
Biochemistry, 45, 2006
|
|
3PLZ
 
 | | Human LRH1 LBD bound to GR470 | | Descriptor: | (3aS,6aR)-5-[(4E)-oct-4-en-4-yl]-N,4-diphenyl-2,3,6,6a-tetrahydropentalen-3a(1H)-amine, 1,2-ETHANEDIOL, FTZ-F1 related protein, ... | | Authors: | Williams, S.P, Xu, R, Zuercher, W.J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small Molecule Agonists of the Orphan Nuclear Receptors Steroidogenic Factor-1 (SF-1, NR5A1) and Liver Receptor Homologue-1 (LRH-1, NR5A2).
J.Med.Chem., 54, 2011
|
|
3O84
 
 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
3O82
 
 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 5'-O-[N-(2,3-dihydroxybenzoyl)sulfamoyl] adenosine | | Descriptor: | 5'-O-{[(2,3-dihydroxyphenyl)carbonyl]sulfamoyl}adenosine, CALCIUM ION, Peptide arylation enzyme | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
2B5J
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with JANSSEN-R165481 | | Descriptor: | (2E)-3-{3-[(5-ETHYL-3-IODO-6-METHYL-2-OXO-1,2-DIHYDROPYRIDIN-4-YL)OXY]PHENYL}ACRYLONITRILE, MANGANESE (II) ION, Reverse transcriptase P51 SUBUNIT, ... | | Authors: | Himmel, D.H, Das, K, Clark Jr, A.D, Hughes, S.H, Benjahad, A, Oumouch, S, Guillemont, J, Coupa, S, Poncelet, A, Csoka, I, Meyer, C, Andries, K, Mguyen, C.H, Grierson, D.S, Arnold, E. | | Deposit date: | 2005-09-28 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures for HIV-1 Reverse Transcriptase in Complexes with Three Pyridinone Derivatives: A New Class of Non-Nucleoside Inhibitors Effective against a Broad Range of Drug-Resistant Strains.
J.Med.Chem., 48, 2005
|
|
2ZNQ
 
 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist TIPP401 | | Descriptor: | (2S)-2-{3-[({[2-fluoro-4-(trifluoromethyl)phenyl]carbonyl}amino)methyl]-4-methoxybenzyl}butanoic acid, Peroxisome proliferator-activated receptor delta, heptyl beta-D-glucopyranoside | | Authors: | Oyama, T, Hirakawa, Y, Nagasawa, N, Miyachi, H, Morikawa, K. | | Deposit date: | 2008-04-30 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1DGI
 
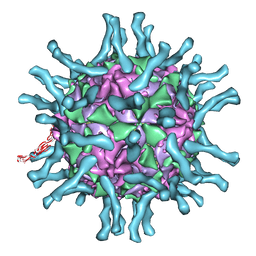 | | Cryo-EM structure of human poliovirus(serotype 1)complexed with three domain CD155 | | Descriptor: | POLIOVIRUS RECEPTOR, VP1, VP2, ... | | Authors: | He, Y, Bowman, V.D, Mueller, S, Bator, C.M, Bella, J, Peng, X, Baker, T.S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 1999-11-24 | | Release date: | 2000-01-24 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (22 Å) | | Cite: | Interaction of the poliovirus receptor with poliovirus.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
