7RJ8
 
 | | CRYSTAL STRUCTURE OF AP2 ASSOCIATED KINASE 1 ISOFORM 1 COMPLEXED WITH LIGAND (2R)-2-AMINO-N-[3-(DIFLUOROM ETHOXY)-4-(1,3-OXAZOL-5-YL)PHENYL]-4-METHYLPENTANAMIDE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 5-[(4-aminopiperidin-1-yl)methyl]-N-{3-[5-(propan-2-yl)-1,3,4-thiadiazol-2-yl]phenyl}pyrrolo[2,1-f][1,2,4]triazin-4-amine, AP2-associated protein kinase 1, ... | | Authors: | Pokross, M, Muckelbauer, J. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Bicyclic Heterocyclic Replacement of an Aryl Amide Leading to Potent and Kinase-Selective Adaptor Protein 2-Associated Kinase 1 Inhibitors.
J.Med.Chem., 65, 2022
|
|
8UH2
 
 | | Complex of C3b with the inhibitor albicin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Albicin, Complement C3 beta chain, ... | | Authors: | Andersen, J.F, Lei, H, Strayer, E, Ribeiro, J.M. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | The mechanism of a mosquito salivary inhibitor of the alternative C3 convertase is elucidated through structural analysis of its complex with C3bBb
To Be Published
|
|
4J9D
 
 | |
8BPC
 
 | | Cryo-EM structure of the human SIN3B histone deacetylase core complex with SAHA at 2.8 Angstrom | | Descriptor: | CALCIUM ION, Histone deacetylase 2, Isoform 2 of Paired amphipathic helix protein Sin3b, ... | | Authors: | Wan, M.S.M, Muhammad, R, Koliopolous, M.G, Alfieri, C. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of assembly, activation and lysine selection by the SIN3B histone deacetylase complex.
Nat Commun, 14, 2023
|
|
4J9F
 
 | |
3GHN
 
 | | Crystal structure of the exosite-containing fragment of human ADAMTS13 (form-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A disintegrin and metalloproteinase with thrombospondin motifs 13, ... | | Authors: | Akiyama, M, Takeda, S, Kokame, K, Takagi, J, Miyata, T. | | Deposit date: | 2009-03-04 | | Release date: | 2009-10-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the non-catalytic domains of ADAMTS13 reveal multiple discontinuous exosites for von Willebrand factor
To be Published
|
|
1I10
 
 | | HUMAN MUSCLE L-LACTATE DEHYDROGENASE M CHAIN, TERNARY COMPLEX WITH NADH AND OXAMATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, L-LACTATE DEHYDROGENASE M CHAIN, ... | | Authors: | Read, J.A, Winter, V.J, Eszes, C.M, Sessions, R.B, Brady, R.L. | | Deposit date: | 2001-01-30 | | Release date: | 2001-03-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase.
Proteins, 43, 2001
|
|
7R0N
 
 | | KRasG12C in complex with GDP and compound 2 | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-02-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C.
Cancer Discov, 12, 2022
|
|
7R0Q
 
 | | KRasG12C in complex with GDP and compound 3 | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-02-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C.
Cancer Discov, 12, 2022
|
|
7R0M
 
 | | KRasG12C in complex with GDP and JDQ443 | | Descriptor: | 1-[6-[4-(5-chloranyl-6-methyl-1~{H}-indazol-4-yl)-5-methyl-3-(1-methylindazol-5-yl)pyrazol-1-yl]-2-azaspiro[3.3]heptan-2-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-02-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C.
Cancer Discov, 12, 2022
|
|
7R26
 
 | | PI3K delta in complex with SD5 | | Descriptor: | 5-[2,6-di(morpholin-4-yl)pyrimidin-4-yl]-4-(trifluoromethyl)pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Gutmann, S, Rummel, G, Shrestha, B. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor.
J.Med.Chem., 65, 2022
|
|
7R2B
 
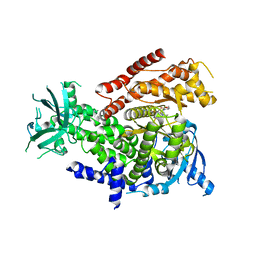 | | PI3Kdelta in complex with an inhibitor | | Descriptor: | (4~{S})-3-[6-[2-azanyl-4-(trifluoromethyl)pyrimidin-5-yl]-2-morpholin-4-yl-pyrimidin-4-yl]-4-methyl-1,3-oxazolidin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Gutmann, S, Rummel, G, Shrestha, B. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor.
J.Med.Chem., 65, 2022
|
|
1JCS
 
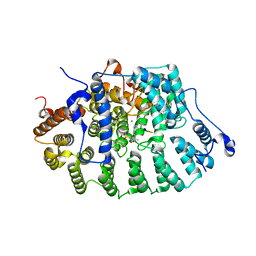 | | CRYSTAL STRUCTURE OF RAT PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH THE PEPTIDE SUBSTRATE TKCVFM AND AN ANALOG OF FARNESYL DIPHOSPHATE | | Descriptor: | ACETIC ACID, PROTEIN FARNESYLTRANSFERASE, ALPHA SUBUNIT, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 2001-06-11 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of human protein farnesyltransferase reveals the basis for inhibition by CaaX tetrapeptides and their mimetics.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
8SUX
 
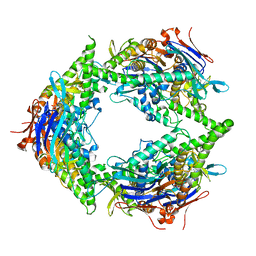 | | Structure of E. coli PtuA hexamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-05-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SXI
 
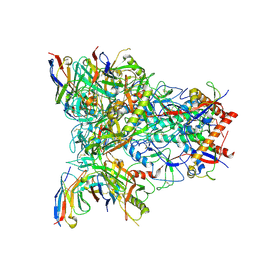 | | CH505 Disulfide Stapled SOSIP Bound to b12 Fab | | Descriptor: | Envelope glycoprotein gp160, HIV-1 gp41, b12 Heavy Chain, ... | | Authors: | Henderson, R. | | Deposit date: | 2023-05-22 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv, 10, 2024
|
|
8R4I
 
 | |
7R1D
 
 | | Structure of MuvB complex | | Descriptor: | Histone-binding protein RBBP4, Protein lin-37 homolog, Protein lin-9 homolog | | Authors: | Koliopoulos, M.G, Alfieri, C. | | Deposit date: | 2022-02-02 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of a nucleosome-bound MuvB transcription factor complex reveals DNA remodelling.
Nat Commun, 13, 2022
|
|
1BN5
 
 | | HUMAN METHIONINE AMINOPEPTIDASE 2 | | Descriptor: | COBALT (II) ION, METHIONINE AMINOPEPTIDASE, TERTIARY-BUTYL ALCOHOL | | Authors: | Liu, S, Widom, J, Kemp, C.W, Crews, C.M, Clardy, J.C. | | Deposit date: | 1998-07-31 | | Release date: | 1999-07-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
1Y9M
 
 | | Crystal structure of exo-inulinase from Aspergillus awamori in spacegroup P212121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Nagem, R.A.P, Rojas, A.L, Golubev, A.M, Korneeva, O.S, Eneyskaya, E.V, Kulminskaya, A.A, Neustroev, K.N, Polikarpov, I. | | Deposit date: | 2004-12-16 | | Release date: | 2004-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of exo-inulinase from Aspergillus awamori: the enzyme fold and structural determinants of substrate recognition
J.Mol.Biol., 344, 2004
|
|
8SXJ
 
 | | CH505 Disulfide Stapled SOSIP Bound to CH235.12 Fab | | Descriptor: | CH235.12 Heavy Chain, CH235.12 Light Chain, Envelope glycoprotein gp160, ... | | Authors: | Henderson, R. | | Deposit date: | 2023-05-22 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv, 10, 2024
|
|
4J9H
 
 | |
8TH1
 
 | |
1CRI
 
 | |
3I97
 
 | | B1 domain of human Neuropilin-1 bound with small molecule EG00229 | | Descriptor: | (S)-2-(3-(benzo[c][1,2,5]thiadiazole-4-sulfonamido)thiophene-2-carboxamido)-5-guanidinopentanoic acid, GLYCEROL, Neuropilin-1 | | Authors: | Allerston, C.K, Djordjevic, S. | | Deposit date: | 2009-07-10 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Small molecule inhibitors of the neuropilin-1 vascular endothelial growth factor A (VEGF-A) interaction.
J.Med.Chem., 53, 2010
|
|
8BPA
 
 | | Cryo-EM structure of the human SIN3B histone deacetylase complex at 3.7 Angstrom | | Descriptor: | CALCIUM ION, Histone deacetylase 2, Isoform 2 of Paired amphipathic helix protein Sin3b, ... | | Authors: | Wan, M.S.M, Muhammad, R, Koliopolous, M.G, Alfieri, C. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of assembly, activation and lysine selection by the SIN3B histone deacetylase complex.
Nat Commun, 14, 2023
|
|
