4D4X
 
 | | Nitrosyl complex of the D121I variant of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C', HEME C, NITRIC OXIDE | | Authors: | Gahfoor, D.D, Kekilli, D, Abdullah, G.H, Dworkowski, F.S.N, Hassan, H.G, Wilson, M.T, Hough, M.A, Strange, R.W. | | Deposit date: | 2014-10-31 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen Bonding of the Dissociated Histidine Ligand is not Required for Formation of a Proximal No Adduct in Cytochrome C'.
J.Biol.Inorg.Chem., 20, 2015
|
|
4KXK
 
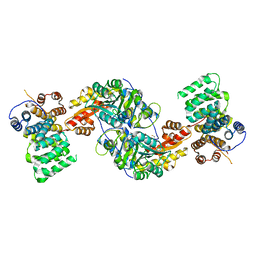 | | Alanine-glyoxylate aminotransferase variant K390A/K391A in complex with the TPR domain of human Pex5p | | Descriptor: | BETA-MERCAPTOETHANOL, Peroxisomal targeting signal 1 receptor, SULFATE ION, ... | | Authors: | Fodor, K, Lou, Y, Wilmanns, M. | | Deposit date: | 2013-05-27 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-Induced Compaction of the PEX5 Receptor-Binding Cavity Impacts Protein Import Efficiency into Peroxisomes.
Traffic, 16, 2015
|
|
4KYO
 
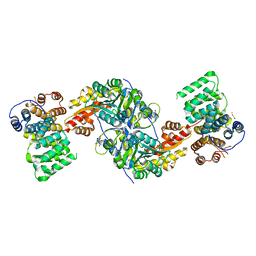 | | Alanine-glyoxylate aminotransferase variant K390A in complex with the TPR domain of human Pex5p | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, Peroxisomal targeting signal 1 receptor, ... | | Authors: | Fodor, K, Lou, Y, Wilmanns, M. | | Deposit date: | 2013-05-29 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-Induced Compaction of the PEX5 Receptor-Binding Cavity Impacts Protein Import Efficiency into Peroxisomes.
Traffic, 16, 2015
|
|
1UC0
 
 | |
6WEN
 
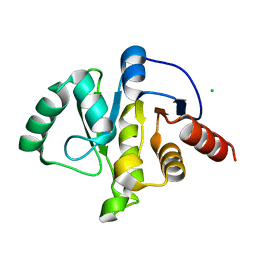 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in the apo form | | Descriptor: | CHLORIDE ION, Non-structural protein 3 | | Authors: | Michalska, K, Stols, L, Jedrzejczak, R, Endres, M, Babnigg, G, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-02 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
1CUI
 
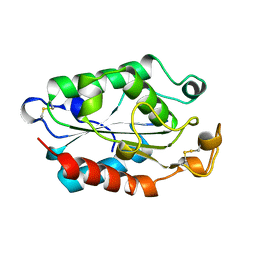 | | CUTINASE, S120A MUTANT | | Descriptor: | CUTINASE | | Authors: | Martinez, C, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
6WT7
 
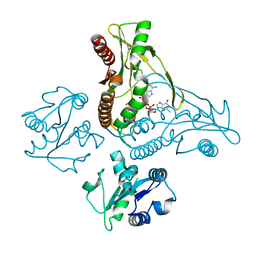 | | Structure of a metazoan TIR-STING receptor from C. gigas in complex with 2',3'-cGAMP | | Descriptor: | Metazoan TIR-STING fusion, cGAMP | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
1CUV
 
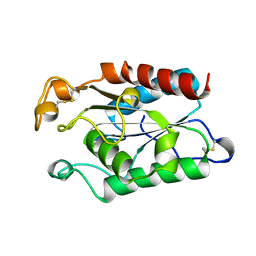 | | CUTINASE, A85F MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUJ
 
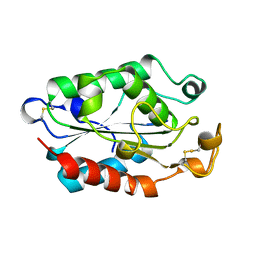 | | CUTINASE, S120C MUTANT | | Descriptor: | CUTINASE | | Authors: | Martinez, C, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
9FB3
 
 | |
1IPG
 
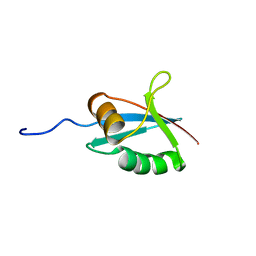 | | SOLUTION STRUCTURE OF THE PB1 DOMAIN OF BEM1P | | Descriptor: | BEM1 PROTEIN | | Authors: | Terasawa, H, Noda, Y, Ito, T, Hatanaka, H, Ichikawa, S, Ogura, K, Sumimoto, H, Inagaki, F. | | Deposit date: | 2001-05-14 | | Release date: | 2001-08-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and ligand recognition of the PB1 domain: a novel protein module binding to the PC motif.
EMBO J., 20, 2001
|
|
9F8U
 
 | |
9F8V
 
 | |
9F8Z
 
 | |
9FDE
 
 | |
1R8T
 
 | | Solution structures of high affinity miniprotein ligands to Streptavidin | | Descriptor: | MP1 | | Authors: | Luo, J, Mukherjee, M, Fan, X, Yang, H, Liu, D, Khan, R, White, M, Fox, R.O. | | Deposit date: | 2003-10-28 | | Release date: | 2005-02-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure-based design of high affinity miniprotein ligands
To be Published
|
|
3OWW
 
 | |
3OXJ
 
 | | crystal structure of glycine riboswitch, soaked in Ba2+ | | Descriptor: | BARIUM ION, GLYCINE, MAGNESIUM ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
9FNV
 
 | |
9FNU
 
 | |
4APA
 
 | |
7YUI
 
 | |
1IEZ
 
 | | Solution Structure of 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase of Riboflavin Biosynthesis | | Descriptor: | 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase | | Authors: | Kelly, M.J.S, Ball, L.J, Kuhne, R, Bacher, A, Oschkinat, H. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the 47-kDa dimeric enzyme 3,4-dihydroxy-2-butanone-4-phosphate synthase and ligand binding studies reveal the location of the active site.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
32C2
 
 | | STRUCTURE OF AN ACTIVITY SUPPRESSING FAB FRAGMENT TO CYTOCHROME P450 AROMATASE | | Descriptor: | IGG1 ANTIBODY 32C2 | | Authors: | Sawicki, M.W, Ng, P.C, Burkhart, B, Pletnev, V, Higashiyama, T, Osawa, Y, Ghosh, D. | | Deposit date: | 1999-04-21 | | Release date: | 2000-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of an activity suppressing Fab fragment to cytochrome P450 aromatase: insights into the antibody-antigen interactions.
Mol.Immunol., 36, 1999
|
|
1WWW
 
 | |
