3L4B
 
 | | Crystal Structure of an Octomeric Two-Subunit TrkA K+ Channel Ring Gating Assembly, TM1088A:TM1088B, from Thermotoga maritima | | Descriptor: | ADENOSINE MONOPHOSPHATE, TrkA K+ Channel protein TM1088A, TrkA K+ Channel protein TM1088B, ... | | Authors: | Deller, M.C, Johnson, H.A, Miller, M, Spraggon, G, Wilson, I.A, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-12-18 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal Structure of an Octomeric Two-Subunit TrkA K+ Channel Ring Gating Assembly, TM1088A:TM1088B, from Thermotoga maritima
To be Published
|
|
4ZDK
 
 | | Crystal structure of the M. tuberculosis CTP synthase PyrG in complex with UTP, AMP-PCP and oxonorleucine | | Descriptor: | 5-OXO-L-NORLEUCINE, CTP synthase, MAGNESIUM ION, ... | | Authors: | Bellinzoni, M, Barilone, N, Alzari, P.M. | | Deposit date: | 2015-04-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Thiophenecarboxamide Derivatives Activated by EthA Kill Mycobacterium tuberculosis by Inhibiting the CTP Synthetase PyrG.
Chem.Biol., 22, 2015
|
|
5AGI
 
 | | Crystal structure of the LeuRS editing domain of Candida albicans Mutant K510A in complex with the adduct formed by AN2690-AMP | | Descriptor: | GLYCEROL, POTENTIAL CYTOSOLIC LEUCYL TRNA SYNTHETASE, [(6-AMINO-9H-PURIN-9-YL)-[5-FLUORO-1,3-DIHYDRO-1-HYDROXY-2,1-BENZOXABOROLE]-4'YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Zhao, H, Palencia, A, Seiradake, E, Ghaemi, Z, Luthey-Schulten, Z, Cusack, S, Martinis, S.A. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Analysis of the Resistance Mechanism of a Benzoxaborole Inhibitor Reveals Insight Into the Leucyl-tRNA Synthetase Editing Mechanism.
Acs Chem.Biol., 10, 2015
|
|
8BMQ
 
 | | Cryo-EM structure of the folate-specific ECF transporter complex in MSP2N2 lipid nanodiscs bound to AMP-PNP | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
5AGJ
 
 | | Crystal structure of the LeuRS editing domain of Candida albicans in complex with the adduct AN2690-AMP | | Descriptor: | POTENTIAL CYTOSOLIC LEUCYL TRNA SYNTHETASE, [(6-AMINO-9H-PURIN-9-YL)-[5-FLUORO-1,3-DIHYDRO-1-HYDROXY-2,1-BENZOXABOROLE]-4'YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Zhao, H, Palencia, A, Seiradake, E, Ghaemi, Z, Luthey-Schulten, Z, Cusack, S, Martinis, S.A. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of the Resistance Mechanism of a Benzoxaborole Inhibitor Reveals Insight Into the Leucyl-tRNA Synthetase Editing Mechanism.
Acs Chem.Biol., 10, 2015
|
|
7RSM
 
 | |
7OZ3
 
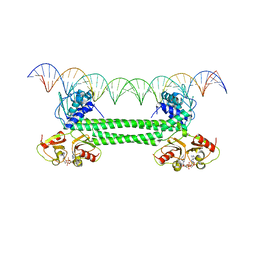 | | S. agalactiae BusR in complex with its busA-promotor DNA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, GntR family transcriptional regulator, pBusA_for, ... | | Authors: | Bandera, A.M, Witte, G. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.46 Å) | | Cite: | BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res., 49, 2021
|
|
5DGH
 
 | |
5UXF
 
 | | Crystal Structure of mouse RECON (AKR1C13) in complex with Cyclic di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Dihydrodiol dehydrogenase | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2017-02-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Sensing of Bacterial Cyclic Dinucleotides by the Oxidoreductase RECON Promotes NF-kappa B Activation and Shapes a Proinflammatory Antibacterial State.
Immunity, 46, 2017
|
|
5EE3
 
 | | COMPLEX STRUCTURE OF OSYCHF1 WITH AMP-PNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1XQZ
 
 | | Crystal Structure of hPim-1 kinase at 2.1 A resolution | | Descriptor: | Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Qian, K.C, Wang, L, Hickey, E.R, Studts, J, Barringer, K, Peng, C, Kronkaitis, A, Li, J, White, A, Mische, S, Farmer, B. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Constitutive Activity and a Unique Nucleotide Binding Mode of Human Pim-1 Kinase.
J.Biol.Chem., 280, 2005
|
|
2A84
 
 | | Crystal structure of A Pantothenate synthetase complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wang, S, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-07 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the Pantothenate Synthetase from Mycobacterium tuberculosis, Snapshots of the Enzyme in Action.
Biochemistry, 45, 2006
|
|
1IVP
 
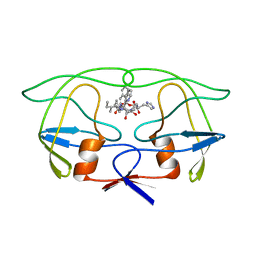 | | THE CRYSTALLOGRAPHIC STRUCTURE OF THE PROTEASE FROM HUMAN IMMUNODEFICIENCY VIRUS TYPE 2 WITH TWO SYNTHETIC PEPTIDIC TRANSITION STATE ANALOG INHIBITORS | | Descriptor: | 4-[(2R)-3-{[(1S,2S,3R,4S)-1-(cyclohexylmethyl)-2,3-dihydroxy-5-methyl-4-({(1S,2R)-2-methyl-1-[(pyridin-2-ylmethyl)carba moyl]butyl}carbamoyl)hexyl]amino}-2-{[(naphthalen-1-yloxy)acetyl]amino}-3-oxopropyl]-1H-imidazol-3-ium, HIV-2 PROTEASE | | Authors: | Mulichak, A.M, Watenpaugh, K.D. | | Deposit date: | 1993-03-18 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystallographic structure of the protease from human immunodeficiency virus type 2 with two synthetic peptidic transition state analog inhibitors.
J.Biol.Chem., 268, 1993
|
|
2XJ0
 
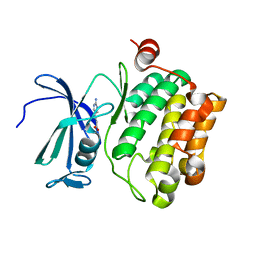 | | Protein kinase Pim-1 in complex with fragment-4 from crystallographic fragment screen | | Descriptor: | (E)-3-(2-AMINO-PYRIDINE-5YL)-ACRYLIC ACID, PROTO-ONCOGENE SERINE/THREONINE PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2EQA
 
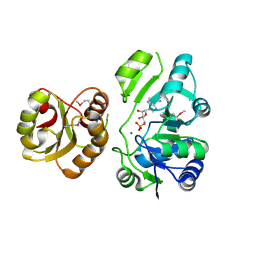 | | Crystal Structure of the hypothetical Sua5 protein from Sulfolobus tokodaii | | Descriptor: | ADENOSINE MONOPHOSPHATE, Hypothetical protein ST1526, MAGNESIUM ION | | Authors: | Agari, Y, Shinkai, A, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of a hypothetical Sua5 protein from Sulfolobus tokodaii strain 7
Proteins, 70, 2008
|
|
2XIX
 
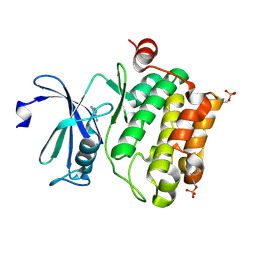 | | Protein kinase Pim-1 in complex with fragment-1 from crystallographic fragment screen | | Descriptor: | 3,5-DIAMINO-1H-[1,2,4]TRIAZOLE, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XJ2
 
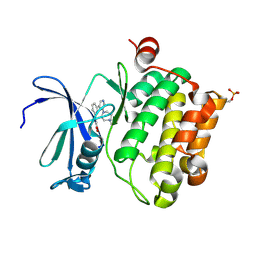 | | Protein kinase Pim-1 in complex with small molecule inhibitor | | Descriptor: | (2E)-3-{3-[6-(4-methyl-1,4-diazepan-1-yl)pyrazin-2-yl]phenyl}prop-2-enoic acid, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XIY
 
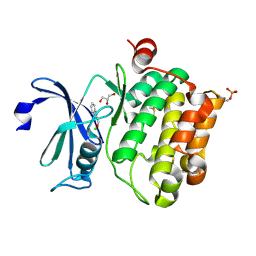 | | Protein kinase Pim-1 in complex with fragment-2 from crystallographic fragment screen | | Descriptor: | 2-HYDROXYMETHYL-BENZOIMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4NDG
 
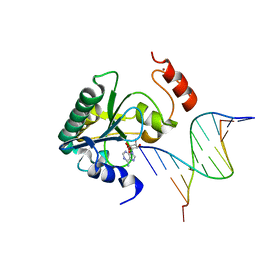 | | Human Aprataxin (Aptx) bound to RNA-DNA and Zn - adenosine vanadate transition state mimic complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
2C38
 
 | | RNase PH core of the archaeal exosome in complex with A5 RNA | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, PROBABLE EXOSOME COMPLEX EXONUCLEASE 1, ... | | Authors: | Lorentzen, E, Conti, E. | | Deposit date: | 2005-10-04 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of 3' End RNA Recognition and Exoribonucleolytic Cleavage by an Exosome Rnase Ph Core.
Mol.Cell, 20, 2005
|
|
2XIZ
 
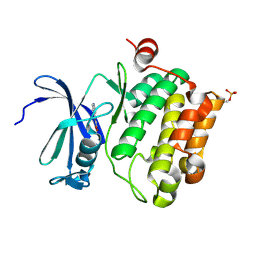 | | Protein kinase Pim-1 in complex with fragment-3 from crystallographic fragment screen | | Descriptor: | (E)-PYRIDIN-4-YL-ACRYLIC ACID, PROTO-ONCOGENE SERINE/THREONINE PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XJ1
 
 | | Protein kinase Pim-1 in complex with small molecule inibitor | | Descriptor: | (2E)-3-(3-{6-[(TRANS-4-AMINOCYCLOHEXYL)AMINO]PYRAZIN-2-YL}PHENYL)PROP-2-ENOIC ACID, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2ZT8
 
 | | Crystal structure of human Glycyl-tRNA synthetase (GlyRS) in complex with Gly-AMP analog | | Descriptor: | Glycyl-tRNA synthetase, [(2S,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (aminoacetyl)sulfamate | | Authors: | Guo, R.T, Yang, X.L, Schimmel, P. | | Deposit date: | 2008-09-19 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structures and biochemical analyses suggest unique mechanism and role for human GlyRS in Ap4A homeostasis
To be Published
|
|
4Q4J
 
 | | Structure of crosslinked TM287/288_S498C_S520C mutant | | Descriptor: | ABC transporter, Uncharacterized ABC transporter ATP-binding protein TM_0288 | | Authors: | Hohl, M, Schoeppe, J, Gruetter, M.G, Seeger, M.A. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4Q4D
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMP-PNP and synthetic 3,5-(PP)2-IP4 (3,5-IP8) | | Descriptor: | (1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 2, MAGNESIUM ION, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2014-04-14 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis of Densely Phosphorylated Bis-1,5-Diphospho-myo-Inositol Tetrakisphosphate and its Enantiomer by Bidirectional P-Anhydride Formation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
