4FNQ
 
 | | Crystal structure of GH36 alpha-galactosidase AgaB from Geobacillus stearothermophilus | | Descriptor: | 1,2-ETHANEDIOL, Alpha-galactosidase AgaB | | Authors: | Merceron, R, Foucault, M, Haser, R, Mattes, R, Watzlawick, H, Gouet, P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular mechanism of the thermostable alpha-galactosidases AgaA and AgaB explained by X-ray crystallography and mutational studies
J.Biol.Chem., 287, 2012
|
|
8RF6
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11A7_AL5 refined against the anomalous diffraction data | | Descriptor: | 6-iodanyl-2,3-dihydro-1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
4FNP
 
 | | Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus | | Descriptor: | Alpha-galactosidase AgaA, SULFATE ION | | Authors: | Merceron, R, Foucault, M, Haser, R, Mattes, R, Watzlawick, H, Gouet, P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | The molecular mechanism of the thermostable alpha-galactosidases AgaA and AgaB explained by X-ray crystallography and mutational studies
J.Biol.Chem., 287, 2012
|
|
4FOL
 
 | | S-formylglutathione hydrolase Variant H160I | | Descriptor: | S-formylglutathione hydrolase | | Authors: | Legler, P.M, Millard, C.B. | | Deposit date: | 2012-06-20 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A role for His-160 in peroxide inhibition of S. cerevisiae S-formylglutathione hydrolase: Evidence for an oxidation sensitive motif.
Arch.Biochem.Biophys., 528, 2012
|
|
8RF5
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11A7 refined against the anomalous diffraction data | | Descriptor: | 6-fluoro-1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
4FQG
 
 | | Crystal structure of the TCERG1 FF4-6 tandem repeat domain | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Transcription elongation regulator 1 | | Authors: | Liu, J, Fan, S, Lee, C.J, Greenleaf, A.L, Zhou, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific Interaction of the Transcription Elongation Regulator TCERG1 with RNA Polymerase II Requires Simultaneous Phosphorylation at Ser2, Ser5, and Ser7 within the Carboxyl-terminal Domain Repeat.
J.Biol.Chem., 288, 2013
|
|
8RF3
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7G3 refined against the anomalous diffraction data | | Descriptor: | 2-(1-benzothiophen-3-yl)ethanoic acid, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
4F9C
 
 | | Human CDC7 kinase in complex with DBF4 and XL413 | | Descriptor: | 8-chloro-2-[(2S)-pyrrolidin-2-yl][1]benzofuro[3,2-d]pyrimidin-4(3H)-one, Cell division cycle 7-related protein kinase, Protein DBF4 homolog A, ... | | Authors: | Hughes, S, Cherepanov, P. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of human CDC7 kinase in complex with its activator DBF4.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FAN
 
 | |
8RCO
 
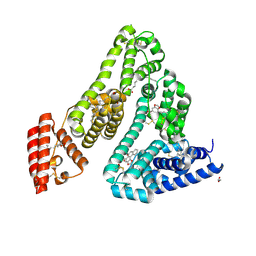 | | Structure of Human Serum Albumin in complex with Aristolochic Acid II at 1.9 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 6-nitronaphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, MYRISTIC ACID, ... | | Authors: | Pomyalov, S, Sidorenko, V.S, Grollman, A.P, Shoham, G. | | Deposit date: | 2023-12-06 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic insights into the transport of aristolochic acids and their active metabolites by human serum albumin.
J.Biol.Chem., 300, 2024
|
|
8RFF
 
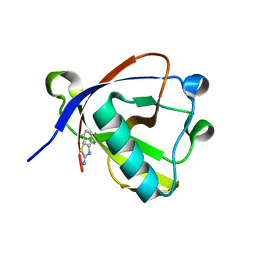 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 6A6 refined against the anomalous diffraction data | | Descriptor: | 1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF4
 
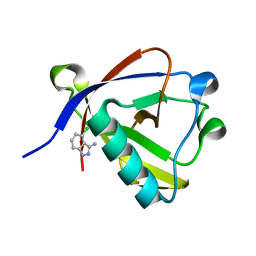 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 9D4 refined against the anomalous diffraction data | | Descriptor: | 4-chloranyl-1~{H}-indazol-3-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF8
 
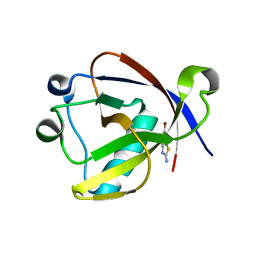 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11A7_AL6 refined against the anomalous diffraction data | | Descriptor: | 6-bromanyl-1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
4FFF
 
 | |
4FF5
 
 | |
8RCP
 
 | | Structure of Human Serum Albumin in complex with Myristic Acid | | Descriptor: | 1,2-ETHANEDIOL, MYRISTIC ACID, Serum albumin | | Authors: | Pomyalov, S, Sidorenko, V.S, Grollman, A.P, Shoham, G. | | Deposit date: | 2023-12-06 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic insights into the transport of aristolochic acids and their active metabolites by human serum albumin.
J.Biol.Chem., 300, 2024
|
|
4FGC
 
 | | Crystal Structure of Active Site Mutant C55A of Nitrile Reductase QueF, Bound to Substrate PreQ0 | | Descriptor: | 2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDINE-5-CARBONITRILE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, ... | | Authors: | Stec, B, Swairjo, M.A. | | Deposit date: | 2012-06-04 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Structural basis of biological nitrile reduction.
J.Biol.Chem., 287, 2012
|
|
4FFS
 
 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Helicobacter pylori with butyl-thio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, CHLORIDE ION, MTA/SAH nucleosidase | | Authors: | Haapalainen, A.M, Rinaldo-Matthis, A, Brown, R.L, Norris, G.E, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-06-01 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Picomolar Transition State Analogue Inhibitor of MTAN as a Specific Antibiotic for Helicobacter pylori.
Biochemistry, 51, 2012
|
|
4FGS
 
 | | Crystal structure of a probable dehydrogenase protein | | Descriptor: | Probable dehydrogenase protein, SULFATE ION | | Authors: | Eswaramoorthy, S, Rice, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-06-04 | | Release date: | 2012-08-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a probable dehydrogenase protein
To be Published
|
|
4FMB
 
 | | VirA-Rab1 complex structure | | Descriptor: | ALUMINUM FLUORIDE, Cysteine protease-like virA, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Shao, F, Zhu, Y. | | Deposit date: | 2012-06-16 | | Release date: | 2012-09-05 | | Last modified: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structurally Distinct Bacterial TBC-like GAPs Link Arf GTPase to Rab1 Inactivation to Counteract Host Defenses.
Cell(Cambridge,Mass.), 150, 2012
|
|
4FNU
 
 | | Crystal structure of GH36 alpha-galactosidase AgaA A355E D478A from Geobacillus stearothermophilus in complex with stachyose | | Descriptor: | Alpha-galactosidase AgaA, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose-(1-6)-alpha-D-galactopyranose-(1-6)-alpha-D-galactopyranose | | Authors: | Merceron, R, Foucault, M, Haser, R, Mattes, R, Watzlawick, H, Gouet, P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The molecular mechanism of the thermostable alpha-galactosidases AgaA and AgaB explained by X-ray crystallography and mutational studies
J.Biol.Chem., 287, 2012
|
|
8T7Z
 
 | |
8SW1
 
 | |
8SW0
 
 | | Puromycin sensitive aminopeptidase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Puromycin-sensitive aminopeptidase, ... | | Authors: | Rodgers, D.W, Sampath, S. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of puromycin-sensitive aminopeptidase and polyglutamine binding.
Plos One, 18, 2023
|
|
2MCY
 
 | | CR1 Sushi domains 2 and 3 | | Descriptor: | Complement receptor type 1 | | Authors: | Park, H.J, Guariento, M.J, Maciejewski, M, Hauart, R, Tham, W, Cowman, A.F, Schmidt, C.Q, Martens, H, Liszewski, K.M, Hourcade, D, Barlow, P.N, Atkinson, J.P. | | Deposit date: | 2013-08-27 | | Release date: | 2013-11-13 | | Last modified: | 2014-01-22 | | Method: | SOLUTION NMR | | Cite: | Using Mutagenesis and Structural Biology to Map the Binding Site for the Plasmodium falciparum Merozoite Protein PfRh4 on the Human Immune Adherence Receptor.
J.Biol.Chem., 289, 2014
|
|
