2YK4
 
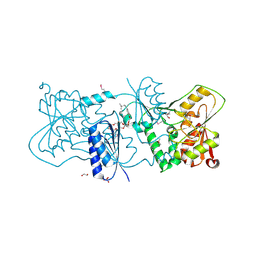 | | Structure of Neisseria LOS-specific sialyltransferase (NST). | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
5XAA
 
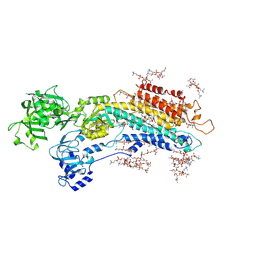 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E2-ALF-(TG) crystals of P21212 symmetry | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
5XA9
 
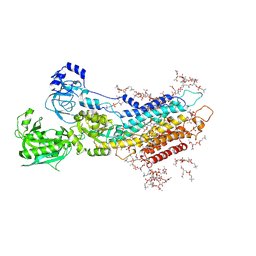 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E2-ALF-(TG) crystals of C2 symmetry | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
5XAB
 
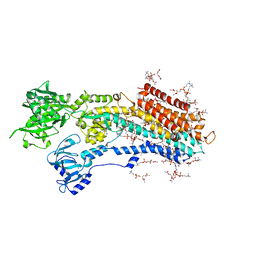 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E2(TG) crystals | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
1GNL
 
 | | Hybrid Cluster Protein from Desulfovibrio desulfuricans X-ray structure at 1.25A resolution using synchrotron radiation at a wavelength of 0.933A | | Descriptor: | ACETATE ION, HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-05 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hybrid Cluster Proteins (Hcps) from Desulfovibrio Desulfuricans Atcc 27774 and Desulfovibrio Vulgaris (Hildenborough): X-Ray Structures at 1.25 A Resolution Using Synchrotron Radiation.
J.Biol.Inorg.Chem., 7, 2002
|
|
6F7T
 
 | |
1GNT
 
 | | Hybrid Cluster Protein from Desulfovibrio vulgaris. X-ray structure at 1.25A resolution using synchrotron radiation. | | Descriptor: | HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hybrid cluster proteins (HCPs) from Desulfovibrio desulfuricans ATCC 27774 and Desulfovibrio vulgaris (Hildenborough): X-ray structures at 1.25 A resolution using synchrotron radiation.
J. Biol. Inorg. Chem., 7, 2002
|
|
4RDL
 
 | | Crystal structure of Norovirus Boxer P domain in complex with Lewis y tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
3OEO
 
 | | The crystal structure E. coli Spy | | Descriptor: | CADMIUM ION, Spheroplast protein Y | | Authors: | Kwon, E, Kim, D.Y, Gross, C.A, Gross, J.D, Kim, K.K. | | Deposit date: | 2010-08-13 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure Escherichia coli Spy.
Protein Sci., 19, 2010
|
|
6FUZ
 
 | | Crystal structure of the TPR domain of KLC1 in complex with the C-terminal peptide of JIP1 | | Descriptor: | GLYCEROL, Kinesin light chain 1,Kinesin light chain 1,C-Jun-amino-terminal kinase-interacting protein 1, nanobody | | Authors: | Pernigo, S, Dodding, M.P, Steiner, R.A. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for isoform-specific kinesin-1 recognition of Y-acidic cargo adaptors.
Elife, 7, 2018
|
|
6FV0
 
 | | Crystal structure of the TPR domain of KLC1 in complex with the C-terminal peptide of torsinA | | Descriptor: | DI(HYDROXYETHYL)ETHER, Kinesin light chain 1,Torsin-1A, nanobody | | Authors: | Pernigo, S, Dodding, M.P, Steiner, R.A. | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis for isoform-specific kinesin-1 recognition of Y-acidic cargo adaptors.
Elife, 7, 2018
|
|
6V2D
 
 | | Crystal Structure of chromodomain of CDYL2 in complex with inhibitor UNC3866 | | Descriptor: | Chromodomain Y-like protein 2, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
3PUN
 
 | | Crystal structure of P domain dimer of Norovirus VA207 with Lewis y tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Chen, Y, Tan, M, Xia, M, Hao, N, Zhang, X.C, Huang, P, Jiang, X, Li, X, Rao, Z. | | Deposit date: | 2010-12-06 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens
Plos Pathog., 7, 2011
|
|
1JX4
 
 | | Crystal Structure of a Y-family DNA Polymerase in a Ternary Complex with DNA Substrates and an Incoming Nucleotide | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-DIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*A)-3', 5'-D(*T*TP*CP*AP*TP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Ling, H, Boudsocq, F, Woodgate, R, Yang, W. | | Deposit date: | 2001-09-05 | | Release date: | 2001-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication.
Cell(Cambridge,Mass.), 107, 2001
|
|
1QFA
 
 | | STRUCTURE OF A NEUROPEPTIDE Y Y2 AGONIST | | Descriptor: | PROTEIN (NEUROPEPTIDE Y) | | Authors: | Barnham, K.J, Catalfamo, F, Pallaghy, P.K, Howlett, G.J, Norton, R.S. | | Deposit date: | 1999-04-08 | | Release date: | 2000-04-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Helical structure and self-association in a 13 residue neuropeptide Y Y2 receptor agonist: relationship to biological activity.
Biochim.Biophys.Acta, 1435, 1999
|
|
1JXL
 
 | | Crystal Structure of a Y-Family DNA Polymerase in a Ternary Complex with DNA Substrates and an Incoming Nucleotide | | Descriptor: | 1,2-ETHANEDIOL, 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*T)-3', ... | | Authors: | Ling, H, Boudsocq, F, Woodgate, R, Yang, W. | | Deposit date: | 2001-09-07 | | Release date: | 2001-10-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication.
Cell(Cambridge,Mass.), 107, 2001
|
|
5T51
 
 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | Descriptor: | KLLA0E05809p, KLLA0F02343p, SULFATE ION | | Authors: | Dimitrova, Y, Jenni, S, Valverde, R, Khin, Y, Harrison, S.C. | | Deposit date: | 2016-08-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2007 Å) | | Cite: | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
4FS1
 
 | | Base pairing mechanism of N2,3-ethenoguanine with dTTP by human polymerase iota | | Descriptor: | DNA 5'-D(*TP*CP*TP*(EFG)P*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*(DOC))-3', DNA polymerase iota, MAGNESIUM ION, ... | | Authors: | Zhao, L. | | Deposit date: | 2012-06-26 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Basis of Miscoding of the DNA Adduct N2,3-Ethenoguanine by Human Y-family DNA Polymerases.
J.Biol.Chem., 287, 2012
|
|
5T58
 
 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | Descriptor: | KLLA0C15939p, KLLA0D15741p, KLLA0E05809p, ... | | Authors: | Dimitrova, Y, Jenni, S, Valverde, R, Khin, Y, Harrison, S.C. | | Deposit date: | 2016-08-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2131 Å) | | Cite: | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
3GIK
 
 | | Dpo4 extension ternary complex with the oxoG(anti)-C(anti) pair | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*AP*AP*CP*(8OG)P*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*(DOC))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-03-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Impact of conformational heterogeneity of OxoG lesions and their pairing partners on bypass fidelity by Y family polymerases.
Structure, 17, 2009
|
|
3GIJ
 
 | | Dpo4 extension ternary complex with oxoG(syn)-A(anti) and oxoG(anti)-A(syn) pairs | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*AP*AP*CP*(8OG)P*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*(2DA))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-03-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Impact of conformational heterogeneity of OxoG lesions and their pairing partners on bypass fidelity by Y family polymerases.
Structure, 17, 2009
|
|
6R2V
 
 | | Arabidopsis NF-Y/CCAAT-box complex | | Descriptor: | FT (-5kb) CCAAT-box 3', FT (-5kb) CCAAT-box 5', NF-YB2, ... | | Authors: | Chaves-Sanjuan, A, Gnesutta, N, Chiara, M, Bernardini, A, Fornara, F, Horner, D, Nardini, M, Mantovani, R. | | Deposit date: | 2019-03-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants.
Plant J., 105, 2021
|
|
3GIM
 
 | | Dpo4 extension ternary complex with oxoG(anti)-G(syn) pair | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*AP*AP*CP*(8OG)P*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*(DDG))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-03-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Impact of conformational heterogeneity of OxoG lesions and their pairing partners on bypass fidelity by Y family polymerases.
Structure, 17, 2009
|
|
5T59
 
 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, KLLA0B13629p, KLLA0E05809p, ... | | Authors: | Dimitrova, Y, Jenni, S, Valverde, R, Khin, Y, Harrison, S.C. | | Deposit date: | 2016-08-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
2AGV
 
 | | Crystal structure of the SR CA2+-ATPASE with BHQ and TG | | Descriptor: | 2,5-DITERT-BUTYLBENZENE-1,4-DIOL, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Toyoshima, C, Obara, K, Norimatsu, Y. | | Deposit date: | 2005-07-27 | | Release date: | 2005-10-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inaugural Article: Structural role of countertransport revealed in Ca2+ pump crystal structure in the absence of Ca2+.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
