2DB3
 
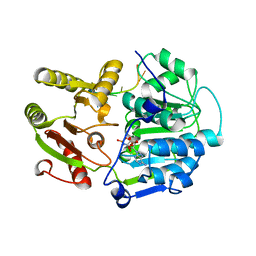 | | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase vasa, MAGNESIUM ION, ... | | Authors: | Sengoku, T, Nureki, O, Nakamura, A, Kobayashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa.
Cell(Cambridge,Mass.), 125, 2006
|
|
7B9V
 
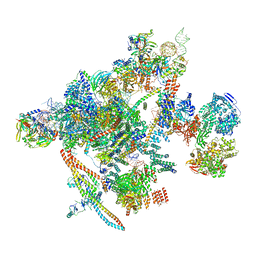 | | Yeast C complex spliceosome at 2.8 Angstrom resolution with Prp18/Slu7 bound | | Descriptor: | 5' exon of UBC4 mRNA, BJ4_G0027490.mRNA.1.CDS.1, BJ4_G0054360.mRNA.1.CDS.1, ... | | Authors: | Wilkinson, M.E, Fica, S.M, Galej, W.P, Nagai, K. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for conformational equilibrium of the catalytic spliceosome.
Mol.Cell, 81, 2021
|
|
5VHA
 
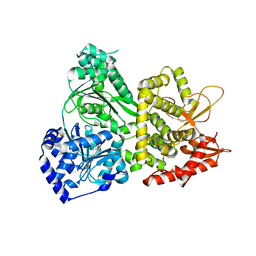 | | DHX36 with an N-terminal truncation | | Descriptor: | DEAH (Asp-Glu-Ala-His) box polypeptide 36 | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
5SUP
 
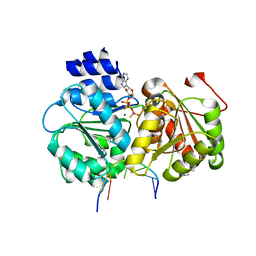 | | Crystal structure of the Sub2-Yra1 complex in association with RNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase SUB2, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ren, Y, Schmiege, P, Blobel, G. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analyses of the DEAD-box ATPase Sub2 in association with THO or Yra1.
Elife, 6, 2017
|
|
7BDK
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8, ... | | Authors: | Vester, K, Santos, K.F, Absmeier, E, Wahl, M.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
J.Biol.Chem., 297, 2021
|
|
7BDJ
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and mant-ATPgammaS | | Descriptor: | MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | Authors: | Vester, K, Santos, K.F, Absmeier, E, Wahl, M.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
J.Biol.Chem., 297, 2021
|
|
7BDI
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and ATPgammaS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Pre-mRNA-processing-splicing factor 8, ... | | Authors: | Vester, K, Santos, K.F, Absmeier, E, Wahl, M.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
J.Biol.Chem., 297, 2021
|
|
7BDL
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and mant-ADP | | Descriptor: | MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | Authors: | Vester, K, Santos, K.F, Absmeier, E, Wahl, M.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
J.Biol.Chem., 297, 2021
|
|
1GM5
 
 | | Structure of RecG bound to three-way DNA junction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-(*CP*AP*GP*CP*TP*CP*CP*AP*TP*GP*AP*TP* CP*AP*TP*TP*GP*GP*CP*A)-3'), DNA (5'-(*GP*AP*GP*CP*AP*CP*TP*GP*C)-3'), ... | | Authors: | Singleton, M.R, Scaife, S, Wigley, D.B. | | Deposit date: | 2001-09-11 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural Analysis of DNA Replication Fork Reversal by Recg
Cell(Cambridge,Mass.), 107, 2001
|
|
1GKU
 
 | |
1OYY
 
 | | Structure of the RecQ Catalytic Core bound to ATP-gamma-S | | Descriptor: | ATP-dependent DNA helicase, MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Bernstein, D.A, Zittel, M.C, Keck, J.L. | | Deposit date: | 2003-04-07 | | Release date: | 2003-10-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-resolution structure of the E. coli RecQ helicase catalytic core
Embo J., 22, 2003
|
|
3RRN
 
 | | S. cerevisiae dbp5 l327v bound to gle1 h337r and ip6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export.
Nature, 472, 2011
|
|
3RRM
 
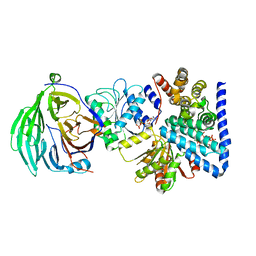 | | S. cerevisiae dbp5 l327v bound to nup159, gle1 h337r, ip6 and adp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export.
Nature, 472, 2011
|
|
1OYW
 
 | |
3SQW
 
 | |
1GL9
 
 | |
1HV8
 
 | |
2EYQ
 
 | |
5SUQ
 
 | | Crystal structure of the THO-Sub2 complex | | Descriptor: | 12-TUNGSTOPHOSPHATE, ATP-dependent RNA helicase SUB2, Tex1, ... | | Authors: | Ren, Y, Blobel, G. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structural and biochemical analyses of the DEAD-box ATPase Sub2 in association with THO or Yra1.
Elife, 6, 2017
|
|
3TBK
 
 | | Mouse RIG-I ATPase Domain | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RIG-I Helicase Domain | | Authors: | Civril, F, Bennett, M.D, Hopfner, K.-P. | | Deposit date: | 2011-08-07 | | Release date: | 2011-10-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling.
Embo Rep., 12, 2011
|
|
3SQX
 
 | |
1FUU
 
 | | YEAST INITIATION FACTOR 4A | | Descriptor: | YEAST INITIATION FACTOR 4A | | Authors: | Caruthers, J.M, Johnson, E.R, McKay, D.B. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5URM
 
 | | Crystal structure of human BRR2 in complex with T-1206548 | | Descriptor: | 3-(5-{[(2R)-5-amino-2-cyclohexyl-7-oxo-2,3-dihydro-7H-[1,3,4]thiadiazolo[3,2-a]pyrimidin-6-yl]methyl}furan-2-yl)benzoic acid, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Klein, M.G, Tjhen, R, Qin, L. | | Deposit date: | 2017-02-11 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
5URJ
 
 | | Crystal structure of human BRR2 in complex with T-3905516 | | Descriptor: | 6-benzyl-3-[(2R)-2-(3-fluoropyridin-2-yl)-6-methyl-3,4-dihydro-2H-1-benzopyran-7-yl]-4,6-dihydropyrido[4,3-d]pyrimidine-2,7(3H,8H)-dione, GLYCEROL, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Klein, M.G, Tjhen, R, Qin, L. | | Deposit date: | 2017-02-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
5URK
 
 | | Crystal structure of human BRR2 in complex with T-3935799 | | Descriptor: | 6-benzyl-3-[3-(benzyloxy)phenyl]-4,6-dihydropyrido[4,3-d]pyrimidine-2,7(1H,3H)-dione, GLYCEROL, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Qin, L, Tjhen, R, Klein, M.G. | | Deposit date: | 2017-02-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
