7K0N
 
 | | Human serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa/ORMDL3, class 2 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Wang, Y, Niu, Y, Zhang, Z, Zhao, H, Myasnikov, A, Kalathur, R, Lee, C.H. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the regulation of human serine palmitoyltransferase complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7K0O
 
 | | Human serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa/ORMDL3, class 3 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Wang, Y, Niu, Y, Zhang, Z, Zhao, H, Myasnikov, A, Kalathur, R, Lee, C.H. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the regulation of human serine palmitoyltransferase complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7K0J
 
 | | Human serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa protomer | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | Authors: | Wang, Y, Niu, Y, Zhang, Z, Zhao, H, Myasnikov, A, Kalathur, R, Lee, C.H. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the regulation of human serine palmitoyltransferase complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7K0Q
 
 | | Human serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa/ORMDL3, myriocin-bound | | Descriptor: | Myriocin, ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Wang, Y, Niu, Y, Zhang, Z, Zhao, H, Myasnikov, A, Kalathur, R, Lee, C.H. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the regulation of human serine palmitoyltransferase complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7K0L
 
 | | Human serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa, myriocin-bound | | Descriptor: | Myriocin, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | Authors: | Wang, Y, Niu, Y, Zhang, Z, Zhao, H, Myasnikov, A, Kalathur, R, Lee, C.H. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the regulation of human serine palmitoyltransferase complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7K0P
 
 | | Human serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa/ORMDL3, class 4 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Wang, Y, Niu, Y, Zhang, Z, Zhao, H, Myasnikov, A, Kalathur, R, Lee, C.H. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the regulation of human serine palmitoyltransferase complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7K0M
 
 | | Human serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa/ORMDL3, class 1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Wang, Y, Niu, Y, Zhang, Z, Zhao, H, Myasnikov, A, Kalathur, R, Lee, C.H. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the regulation of human serine palmitoyltransferase complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7K0I
 
 | | Human serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | Authors: | Wang, Y, Niu, Y, Zhang, Z, Zhao, H, Myasnikov, A, Kalathur, R, Lee, C.H. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the regulation of human serine palmitoyltransferase complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7CQI
 
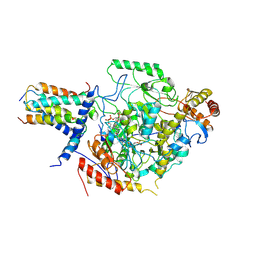 | | Cryo-EM structure of the substrate-bound SPT-ORMDL3 complex | | Descriptor: | ORM1-like protein 3, Serine palmitoyltransferase 1, Serine palmitoyltransferase 2, ... | | Authors: | Li, S.S, Xie, T, Wang, L, Gong, X. | | Deposit date: | 2020-08-11 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7CQK
 
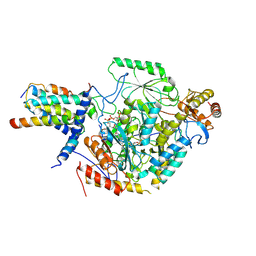 | | Cryo-EM structure of the substrate-bound SPT-ORMDL3 complex | | Descriptor: | ORM1-like protein 3, Serine palmitoyltransferase 1, Serine palmitoyltransferase 2, ... | | Authors: | Li, S.S, Xie, T, Wang, L, Gong, X. | | Deposit date: | 2020-08-11 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZVG
 
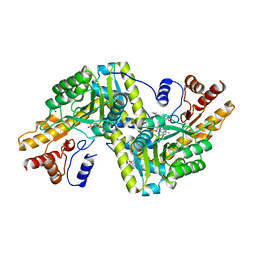 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-indole-3-lactic acid | | Descriptor: | 3-(INDOL-3-YL) LACTATE, Aminotransferase, MAGNESIUM ION, ... | | Authors: | Rum, J, Rutkiewicz, M, Pruska, A, Bujacz, A, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | Deposit date: | 2020-07-24 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
6ZUP
 
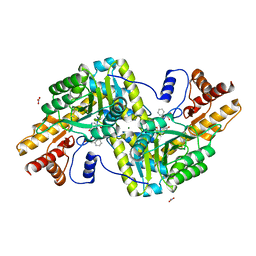 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-(-)-3-phenyllactic acid | | Descriptor: | ALPHA-HYDROXY-BETA-PHENYL-PROPIONIC ACID, Aminotransferase, MAGNESIUM ION, ... | | Authors: | Bujacz, A, Rum, J, Rutkiewicz, M, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
6ZUR
 
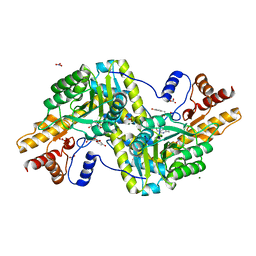 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-p-hydroxyphenyllactic acid | | Descriptor: | (2S)-2-hydroxy-3-(4-hydroxyphenyl)propanoic acid, Aminotransferase, MAGNESIUM ION, ... | | Authors: | Bujacz, A, Rum, J, Rutkiewicz, M, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
7BXS
 
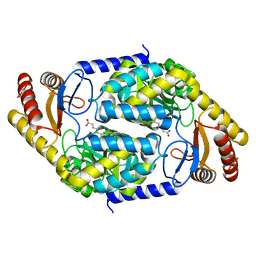 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator glycine binding form | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
7BXQ
 
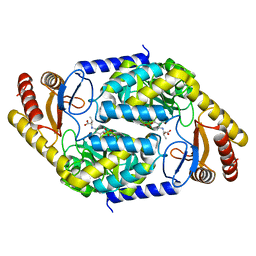 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator L-Threonine binding form | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-allothreonine | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
7BXP
 
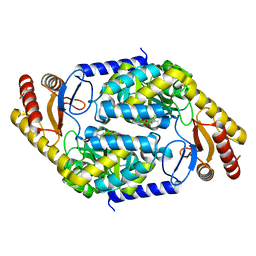 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
7BXR
 
 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator 3-Hydroxynorvaline binding form | | Descriptor: | (2S,3R)-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-3-oxidanyl-pentanoic acid, 2-amino-3-ketobutyrate coenzyme A ligase | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
6M4O
 
 | | Cryo-EM structure of the monomeric SPT-ORMDL3 complex | | Descriptor: | ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | Authors: | Li, S.S, Xie, T, Wang, L, Gong, X. | | Deposit date: | 2020-03-08 | | Release date: | 2021-02-10 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6M4N
 
 | | Cryo-EM structure of the dimeric SPT-ORMDL3 complex | | Descriptor: | ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | Authors: | Li, S.S, Xie, T, Wang, L, Gong, X. | | Deposit date: | 2020-03-07 | | Release date: | 2021-02-10 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6LIG
 
 | |
6UXZ
 
 | | (S)-4-Amino-5-phenoxypentanoate as a Selective Agonist of the Transcription Factor GabR | | Descriptor: | (4S)-4-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-5-phenoxypentanoic acid, HTH-type transcriptional regulatory protein GabR, SULFATE ION | | Authors: | Catlin, D.S, Liu, D. | | Deposit date: | 2019-11-09 | | Release date: | 2020-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | (S)-4-Amino-5-phenoxypentanoate designed as a potential selective agonist of the bacterial transcription factor GabR.
Protein Sci., 29, 2020
|
|
6T8P
 
 | | HKATII IN COMPLEX WITH LIGAND (2R)-N-benzyl-1-[6-methyl-5-(oxan-4-yl)-7-oxo-6H,7H-[1,3]thiazolo[5,4-d]pyrimidin-2-yl]pyrrolidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 3,5-bis(fluoranyl)-~{N}-[5-[(2~{R})-2-(3-fluorophenyl)-3-methyl-butyl]-1,3,4-thiadiazol-2-yl]benzenesulfonamide, IODIDE ION, ... | | Authors: | Blaesse, M, Venalainen, J. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery of sulfonamides and 9-oxo-2,8-diazaspiro[5,5]undecane-2-carboxamides as human kynurenine aminotransferase 2 (KAT2) inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6T8Q
 
 | | HKATII IN COMPLEX WITH LIGAND (2R)-N-benzyl-1-[6-methyl-5-(oxan-4-yl)-7-oxo-6H,7H-[1,3]thiazolo[5,4-d]pyrimidin-2-yl]pyrrolidine-2-carboxamide | | Descriptor: | (2~{R})-1-[6-methyl-5-(oxan-4-yl)-7-oxidanylidene-[1,3]thiazolo[5,4-d]pyrimidin-2-yl]-~{N}-(phenylmethyl)pyrrolidine-2-carboxamide, ACETATE ION, CADMIUM ION, ... | | Authors: | Blaesse, M, Venalainen, J. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of sulfonamides and 9-oxo-2,8-diazaspiro[5,5]undecane-2-carboxamides as human kynurenine aminotransferase 2 (KAT2) inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6T3V
 
 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - malic acid | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rutkiewicz, M, Bujacz, A, Rum, J, Bujacz, G. | | Deposit date: | 2019-10-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials, 14, 2021
|
|
6L1N
 
 | |
