5Y97
 
 | | Crystal structure of snake gourd seed lectin in complex with lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Seed lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Chandran, T, Vijayan, M, Sivaji, N, Surolia, A. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Ligand binding and retention in snake gourd seed lectin (SGSL). A crystallographic, thermodynamic and molecular dynamics study
Glycobiology, 28, 2018
|
|
5Y42
 
 | |
2G81
 
 | | Crystal Structure of the Bowman-Birk Inhibitor from Vigna unguiculata Seeds in Complex with Beta-trypsin at 1.55 Angstrons Resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Bowman-Birk type seed trypsin and chymotrypsin inhibitor, ... | | Authors: | Freitas, S.M, Barbosa, J.A.R.G, Paulino, L.S, Teles, R.C.L, Esteves, G.F, Ventura, M.M. | | Deposit date: | 2006-03-01 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the Bowman-Birk Inhibitor from Vigna unguiculata Seeds in Complex with {beta}-Trypsin at 1.55 A Resolution and Its Structural Properties in Association with Proteinases
Biophys.J., 92, 2007
|
|
7NCJ
 
 | |
7NCG
 
 | |
7NCI
 
 | |
7NCH
 
 | |
7NCK
 
 | |
7NCA
 
 | |
6STQ
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with N,N'-Diacetylchitobiose; 30 seconds soaking | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Arundo donax Lectin (ADL), GLYCEROL | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STM
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) | | Descriptor: | Arundo donax Lectin (ADL), GLYCEROL | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STP
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with sialic acid | | Descriptor: | Arundo donax Lectin (ADL), GLYCEROL, N-acetyl-alpha-neuraminic acid | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STO
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with N-Acetyl lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Arundo donax Lectin (ADL), GLYCEROL, ... | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
4L8Q
 
 | | Crystal structure of Canavalia grandiflora seed lectin complexed with X-Man. | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CADMIUM ION, CALCIUM ION, ... | | Authors: | Barroso-Neto, I.L, Rocha, B.A.M, Simoes, R.C, Bezerra, M.J.B, Pereira-Junior, F.N, Osterne, V.J.S, Nascimento, K.S, Nagano, C.S, Delatorre, P, Sampaio, A.H, Cavada, B.S. | | Deposit date: | 2013-06-17 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Vasorelaxant activity of Canavalia grandiflora seed lectin: A structural analysis.
Arch.Biochem.Biophys., 543, 2014
|
|
1H9W
 
 | | Native Dioclea Guianensis seed lectin | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, SEED LECTIN | | Authors: | Romero, A, Wah, D.A, Sol, F.G.D, Cavada, B.S, Ramos, M.V, Grangeiro, T.B, Sampaio, A.H, Calvete, J.J. | | Deposit date: | 2001-03-22 | | Release date: | 2001-03-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Native and Cd/Cd-Substituted Dioclea Guianensis Seed Lectin. A Novel Manganese-Binding Site and Structural Basis of Dimer-Tetramer Association
J.Mol.Biol., 310, 2001
|
|
1JG5
 
 | | CRYSTAL STRUCTURE OF RAT GTP CYCLOHYDROLASE I FEEDBACK REGULATORY PROTEIN, GFRP | | Descriptor: | GTP CYCLOHYDROLASE I FEEDBACK REGULATORY PROTEIN, POTASSIUM ION | | Authors: | Bader, G, Schiffmann, S, Herrmann, A, Fischer, M, Gutlich, M, Auerbach, G, Ploom, T, Bacher, A, Huber, R, Lemm, T. | | Deposit date: | 2001-06-23 | | Release date: | 2001-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of rat GTP cyclohydrolase I feedback regulatory protein, GFRP.
J.Mol.Biol., 312, 2001
|
|
2R33
 
 | |
4X3E
 
 | |
6V3X
 
 | |
6V3Y
 
 | |
6U4Y
 
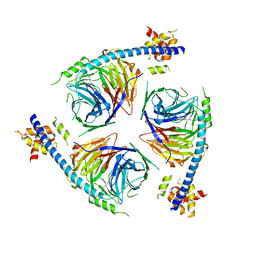 | | Crystal Structure of an EZH2-EED Complex in an Oligomeric State | | Descriptor: | Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Jiao, L, Liu, X. | | Deposit date: | 2019-08-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A partially disordered region connects gene repression and activation functions of EZH2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5WP3
 
 | | Crystal Structure of EED in complex with EB22 | | Descriptor: | EB22, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Zhu, L, Moody, J.D, Baker, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | First critical repressive H3K27me3 marks in embryonic stem cells identified using designed protein inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3K27
 
 | | Complex structure of EED and trimethylated H3K9 | | Descriptor: | HISTONE PEPTIDE, Polycomb protein EED | | Authors: | Bian, C.B, Xu, C, Qiu, W, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-29 | | Release date: | 2009-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JZG
 
 | | Structure of EED in complex with H3K27me3 | | Descriptor: | HISTONE PEPTIDE, Polycomb protein EED | | Authors: | Xu, C, Bian, C.B, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-23 | | Release date: | 2009-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3K26
 
 | | Complex structure of EED and trimethylated H3K4 | | Descriptor: | HISTONE PEPTIDE, Polycomb protein EED | | Authors: | Bian, C.B, Xu, C, Qiu, W, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-29 | | Release date: | 2009-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
