4XV0
 
 | | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Trichoderma reesei | | Descriptor: | Beta-xylanase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Savchenko, A. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9697 Å) | | Cite: | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Trichoderma reesei
To Be Published
|
|
6ODC
 
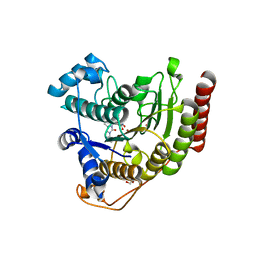 | | Crystal structure of HDAC8 in complex with compound 30 | | Descriptor: | (2E)-3-[2-(3-cyclopentyl-5,5-dimethyl-2-oxoimidazolidin-1-yl)phenyl]-N-hydroxyprop-2-enamide, 1,2-ETHANEDIOL, Histone deacetylase 8, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
4XH6
 
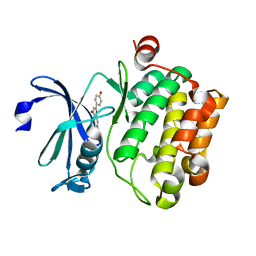 | | Crystal structure of proto-oncogene kinase Pim1 bound to hispidulin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-6-methoxy-4H-chromen-4-one, Serine/threonine-protein kinase pim-1 | | Authors: | Su, M.-Y, Chang, C.-I. | | Deposit date: | 2015-01-05 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Total Synthesis of Hispidulin and the Structural Basis for Its Inhibition of Proto-oncogene Kinase Pim-1
J.Nat.Prod., 78, 2015
|
|
6WFM
 
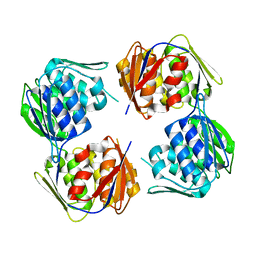 | |
6OKO
 
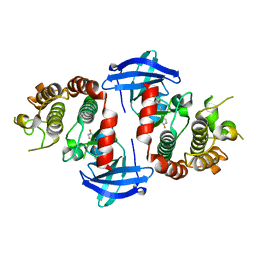 | |
6OI9
 
 | | Crystal Structure of E. coli Biotin Carboxylase Complexed with 7-[3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 7-[(3S)-3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine, Biotin carboxylase | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Optimization and Mechanistic Characterization of Pyridopyrimidine Inhibitors of Bacterial Biotin Carboxylase.
J.Med.Chem., 62, 2019
|
|
7PPZ
 
 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) C94S inactive mutant in complex with human eIF4A - Crystal form A | | Descriptor: | Burkholderia Lethal Factor 1 (BLF1), Eukaryotic initiation factor 4A-I | | Authors: | Mobbs, G.W, Aziz, A.A, Dix, S.R, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2021-09-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
6OAZ
 
 | | Apo Structure of WT Lipoprotein Lipase in Complex with GPIHBP1 Mutant N78D N82D produced in HEK293-F cells | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Arora, R, Horton, P.A, Benson, T.E, Romanowski, M.J. | | Deposit date: | 2019-03-19 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of lipoprotein lipase in complex with GPIHBP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4Y2J
 
 | |
8VA6
 
 | | Menin in complex with Ziftomenib (KO-539) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Clegg, B.D, Linhares, B.M, Cierpicki, T, Grembecka, J. | | Deposit date: | 2023-12-11 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Ziftomenib in relapsed or refractory acute myeloid leukaemia (KOMET-001): a multicentre, open-label, multi-cohort, phase 1 trial.
Lancet Oncol, 25, 2024
|
|
4XYK
 
 | | Crystal structure of human phosphofructokinase-1 in complex with ADP, Northeast Structural Genomics Consortium Target HR9275 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent 6-phosphofructokinase, platelet type, ... | | Authors: | Forouhar, F, Webb, B.A, Szu, F.-E, Seetharaman, J, Barber, D.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations.
Nature, 523, 2015
|
|
6XE6
 
 | | Structure of Human Dispatched-1 (DISP1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1 | | Authors: | Chen, H, Liu, Y, Li, X. | | Deposit date: | 2020-06-12 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structure of human Dispatched-1 provides insights into Hedgehog ligand biogenesis.
Life Sci Alliance, 3, 2020
|
|
6QU4
 
 | | Crystal Structure of Phosphofructokinase from Trypanosoma brucei in complex with an allosteric inhibitor ctcb405 | | Descriptor: | 1-[(3,4-dichlorophenyl)methyl]-5-[2-(dimethylamino)ethyl]pyrrolo[3,2-c]pyridin-4-one, ATP-dependent 6-phosphofructokinase, CITRIC ACID, ... | | Authors: | McNae, I.W, Dornan, J, Walkinshaw, M.D. | | Deposit date: | 2019-02-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Fast acting allosteric phosphofructokinase inhibitors block trypanosome glycolysis and cure acute African trypanosomiasis in mice.
Nat Commun, 12, 2021
|
|
7PX5
 
 | | ATAD2 in complex with 1-Methyl-2-quinolone | | Descriptor: | 1,2-ETHANEDIOL, 1-METHYLQUINOLIN-2(1H)-ONE, ATPase family AAA domain-containing protein 2, ... | | Authors: | Martin, M.P, Noble, M.E.N. | | Deposit date: | 2021-10-07 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Exiting the tunnel of uncertainty: crystal soak to validated hit.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8VA5
 
 | | Menin mutant - T349M in complex with Ziftomenib (KO-539) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Clegg, B.D, Cierpicki, T, Grembecka, J. | | Deposit date: | 2023-12-11 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Ziftomenib in relapsed or refractory acute myeloid leukaemia (KOMET-001): a multicentre, open-label, multi-cohort, phase 1 trial.
Lancet Oncol, 25, 2024
|
|
6P74
 
 | | OLD nuclease from Thermus Scotoductus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PLATINUM (II) ION, Putative ATP-dependent endonuclease of the OLD family, ... | | Authors: | Chappie, J.S, Schiltz, C.J. | | Deposit date: | 2019-06-04 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The full-length structure of Thermus scotoductus OLD defines the ATP hydrolysis properties and catalytic mechanism of Class 1 OLD family nucleases.
Nucleic Acids Res., 48, 2020
|
|
9DE6
 
 | | Structure of full-length HIV TAR RNA | | Descriptor: | MAGNESIUM ION, RNA (57-MER) | | Authors: | Bou-Nader, C, Zhang, J. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of complete HIV-1 TAR RNA portray a dynamic platform poised for protein binding and structural remodeling.
Nat Commun, 16, 2025
|
|
8V2L
 
 | | Crystal structure of IRAK4 kinase domain with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-1 receptor-associated kinase 4, N-{2-[4-(hydroxymethyl)phenyl]-6-(2-hydroxypropan-2-yl)-2H-indazol-5-yl}-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8ZXX
 
 | | PfDXR - Mn2+ - NADPH - TAKK442 quaternary complex | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, GLYCEROL, ... | | Authors: | Tanaka, N, Takada, S, Sakamoto, Y. | | Deposit date: | 2024-06-15 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Expanding the Chemical Space of Reverse Fosmidomycin Analogs.
Acs Med.Chem.Lett., 16, 2025
|
|
5YL9
 
 | |
8X4O
 
 | |
7CF6
 
 | | Crystal structure of Beta-aspartyl dipeptidase from thermophilic keratin degrading Fervidobacterium islandicum AW-1 in complex with beta-Asp-Leu dipeptide | | Descriptor: | (2S)-2-[[(3S)-3-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]amino]-4-methyl-pentanoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Dhanasingh, I, La, J.W, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Functional Characterization of Primordial Protein Repair Enzyme M38 Metallo-Peptidase From Fervidobacterium islandicum AW-1.
Front Mol Biosci, 7, 2020
|
|
1PG9
 
 | | NMR Solution Structure of an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link in a DNA Dodecamer Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3', CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II) | | Authors: | Wu, Y, Pradhan, P, Havener, J, Chaney, S.G, Campbel, S.L. | | Deposit date: | 2003-05-28 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oxaliplatin 1,2-d(GG) intrastrand cross-link in a DNA dodecamer duplex
J.Mol.Biol., 341, 2004
|
|
9HR9
 
 | |
4WDI
 
 | |
