5KVY
 
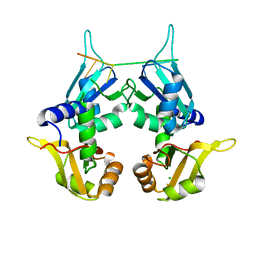 | | CRYSTAL STRUCTURE OF THE TWO TANDEM RRM DOMAINS OF PUF60 BOUND TO A PORTION OF AN ADML PRE-MRNA 3' SPLICE SITE ANALOG | | Descriptor: | CHLORIDE ION, DNA (30-MER), Poly(U)-binding-splicing factor PUF60 | | Authors: | Hsiao, H.-H, Crichlow, G.V, Albright, R.A, Murphy, J.W, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-15 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
7EPG
 
 | |
7EPI
 
 | |
7EPJ
 
 | |
5KW6
 
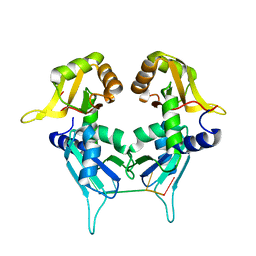 | | Two Tandem RRM Domains of PUF60 Bound to an AdML Pre-mRNA 3' Splice Site Analogue with a Modified Binding-Site Nucleic Acid Base | | Descriptor: | DNA (30-MER), Poly(U)-binding-splicing factor PUF60 | | Authors: | Crichlow, G.V, Hsiao, H.-H, Albright, R, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-15 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
7NSQ
 
 | |
7NSO
 
 | |
7NSP
 
 | |
6VZJ
 
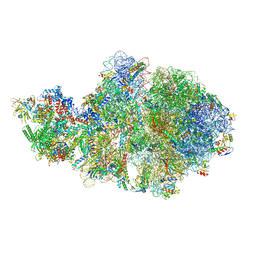 | | Escherichia coli transcription-translation complex A1 (TTC-A1) containing mRNA with a 15 nt long spacer, fMet-tRNAs at E-site and P-site, and lacking transcription factor NusG | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-28 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
5LOE
 
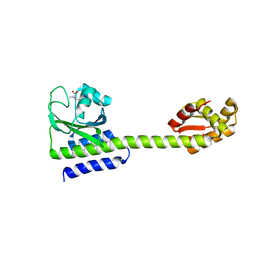 | | Structure of full length Cody from Bacillus subtilis in complex with Ile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, ISOLEUCINE | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2016-08-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Branched-chain Amino Acid and GTP-sensing Global Regulator, CodY, from Bacillus subtilis.
J. Biol. Chem., 292, 2017
|
|
5LOJ
 
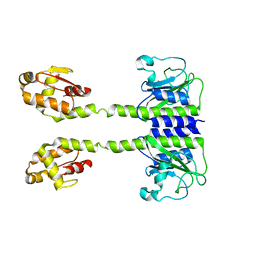 | |
5LNH
 
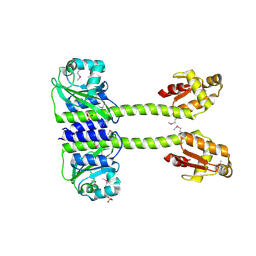 | | Structure of full length Unliganded CodY from Bacillus subtilis | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, SULFATE ION | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2016-08-04 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Branched-chain Amino Acid and GTP-sensing Global Regulator, CodY, from Bacillus subtilis.
J. Biol. Chem., 292, 2017
|
|
8C78
 
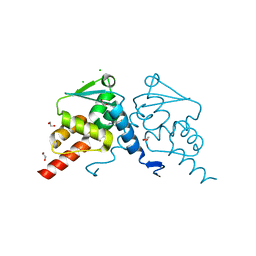 | | Crystal structure of human BCL6 BTB domain in complex with compound CCT374705 | | Descriptor: | (2~{S})-10-[(3-chloranyl-2-fluoranyl-pyridin-4-yl)amino]-2-cyclopropyl-3,3-bis(fluoranyl)-7-methyl-2,4-dihydro-1~{H}-[1,4]oxazepino[2,3-c]quinolin-6-one, 1,2-ETHANEDIOL, B-cell lymphoma 6 protein, ... | | Authors: | Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an In Vivo Chemical Probe for BCL6 Inhibition by Optimization of Tricyclic Quinolinones.
J.Med.Chem., 66, 2023
|
|
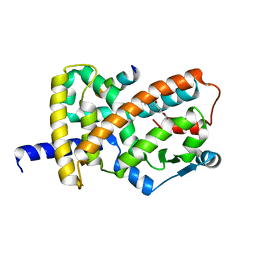
6ONI
 
 | |
5YDV
 
 | | Regulatory domain of HypT from Salmonella typhimurium complexed with HOCl (HOCl-bound form) | | Descriptor: | Cell density-dependent motility repressor, SULFATE ION, hypochlorous acid | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-14 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5YEZ
 
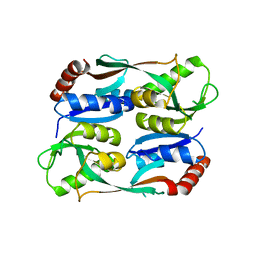 | | Regulatory domain of HypT M206Q mutant from Salmonella typhimurium | | Descriptor: | Cell density-dependent motility repressor | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-20 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5YDW
 
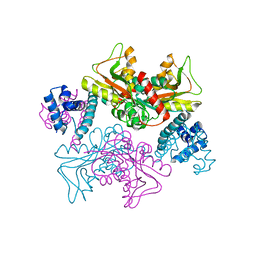 | | Full-length structure of HypT from Salmonella typhimuriuma (hypochlorite-specific LysR-type transcriptional regulator) | | Descriptor: | Cell density-dependent motility repressor | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5YER
 
 | | Regulatory domain of HypT from Salmonella typhimurium (Bromide ion-bound) | | Descriptor: | BROMIDE ION, Cell density-dependent motility repressor, SULFATE ION | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-19 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5Z28
 
 | | AtVAL2 PHD-Like domain | | Descriptor: | B3 domain-containing transcription repressor VAL2, GLYCEROL, ZINC ION | | Authors: | Wu, B.X, Zhang, M.M. | | Deposit date: | 2017-12-30 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Crystal structure of AtVAL12 PHD-Like domain
To Be Published
|
|
6A2S
 
 | | Mycobacterium tuberculosis LexA C-domain S160A | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, LexA repressor | | Authors: | Chandran, A.V, Srikalaivani, R, Paul, A, Vijayan, M. | | Deposit date: | 2018-06-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical characterization of Mycobacterium tuberculosis LexA and structural studies of its C-terminal segment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6A2Q
 
 | | Mycobacterium tuberculosis LexA C-domain I | | Descriptor: | GLYCEROL, LexA repressor | | Authors: | Chandran, A.V, Srikalaivani, R, Paul, A, Vijayan, M. | | Deposit date: | 2018-06-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Biochemical characterization of Mycobacterium tuberculosis LexA and structural studies of its C-terminal segment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6A2R
 
 | | Mycobacterium tuberculosis LexA C-domain II | | Descriptor: | DI(HYDROXYETHYL)ETHER, LexA repressor | | Authors: | Chandran, A.V, Srikalaivani, R, Paul, A, Vijayan, M. | | Deposit date: | 2018-06-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Biochemical characterization of Mycobacterium tuberculosis LexA and structural studies of its C-terminal segment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6A2T
 
 | | Mycobacterium tuberculosis LexA C-domain K197A | | Descriptor: | ACRYLIC ACID, LexA repressor | | Authors: | Chandran, A.V, Srikalaivani, R, Paul, A, Vijayan, M. | | Deposit date: | 2018-06-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical characterization of Mycobacterium tuberculosis LexA and structural studies of its C-terminal segment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4YZ6
 
 | | Crystal Structure of Myc3[44-238] from Arabidopsis in complex with Jaz1 peptide [200-221] | | Descriptor: | Protein TIFY 10A, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J, Xu, H.E, Melcher, K, HE, S.Y. | | Deposit date: | 2015-03-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
6KTB
 
 | | Crystal structure of B. halodurans MntR in apo form | | Descriptor: | HTH-type transcriptional regulator MntR, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Lee, J.Y, Lee, M.Y. | | Deposit date: | 2019-08-26 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the manganese transport regulator MntR from Bacillus halodurans in apo and manganese bound forms.
Plos One, 14, 2019
|
|
