7S85
 
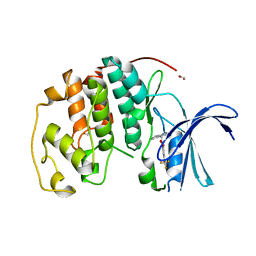 | | Crystal structure of CDK2 liganded with compound WN316 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(trifluoromethoxy)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7N11
 
 | |
7PTL
 
 | |
5MQJ
 
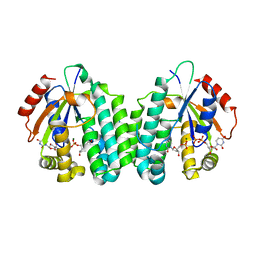 | | Crystal structure of dCK mutant C3S | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Deoxycytidine kinase, MAGNESIUM ION, ... | | Authors: | Saez-Ayala, M, Rebuffet, E, Hammam, K, Gros, L, Lopez, S, Hajem, B, Humbert, M, Baudelet, E, Audebert, S, Betzi, S, Lugari, A, Combes, S, Pez, D, Letard, S, Mansfield, C, Moussy, A, de Sepulveda, P, Morelli, X, Dubreuil, P. | | Deposit date: | 2016-12-20 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Dual protein kinase and nucleoside kinase modulators for rationally designed polypharmacology.
Nat Commun, 8, 2017
|
|
5MQW
 
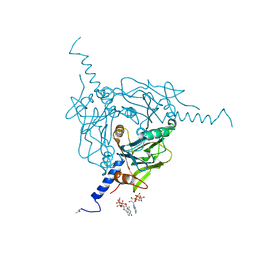 | | High-speed fixed-target serial virus crystallography | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Roedig, P, Ginn, H.M, Pakendorf, T, Sutton, G, Harlos, K, Walter, T.S, Meyer, J, Fischer, P, Duman, R, Vartiainen, I, Reime, B, Warmer, M, Brewster, A.S, Young, I.D, Michels-Clark, T, Sauter, N.K, Sikorsky, M, Nelson, S, Damiani, D.S, Alonso-Mori, R, Ren, J, Fry, E.E, David, C, Stuart, D.I, Wagner, A, Meents, A. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-speed fixed-target serial virus crystallography.
Nat. Methods, 14, 2017
|
|
7PTK
 
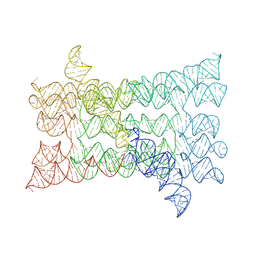 | |
5TXN
 
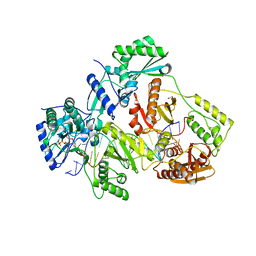 | | STRUCTURE OF Q151M MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DATP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | Authors: | Das, K, Martinez, S.M, Arnold, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
8THD
 
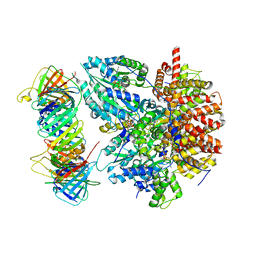 | | Structure of the Saccharomyces cerevisiae clamp unloader Elg1-RFC bound to PCNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ELG1 isoform 1, MAGNESIUM ION, ... | | Authors: | Zheng, F, Yao, Y.N, Georgescu, R, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-07-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structure of the PCNA unloader Elg1-RFC.
Sci Adv, 10, 2024
|
|
8THB
 
 | | Structure of the Saccharomyces cerevisiae PCNA clamp unloader Elg1-RFC complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ELG1 isoform 1, MAGNESIUM ION, ... | | Authors: | Zheng, F, Yao, Y.N, Georgescu, R, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-07-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the PCNA unloader Elg1-RFC.
Sci Adv, 10, 2024
|
|
1FFN
 
 | | CRYSTAL STRUCTURE OF MURINE CLASS I H-2DB COMPLEXED WITH PEPTIDE GP33(C9M) | | Descriptor: | BETA-2 MICROGLOBULIN, BETA CHAIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Wang, B, Sharma, A, Maile, R, Saad, M, Collins, E.J, Frelinger, J.A. | | Deposit date: | 2000-07-25 | | Release date: | 2002-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Peptidic termini play a significant role in TCR recognition
J.IMMUNOL., 169, 2002
|
|
6XSD
 
 | | Patient-derived B2GPI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1 | | Authors: | Klenotic, P.A, Yu, E.W.Y. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | B2-Glycoprotein I and it's role in APS
To Be Published
|
|
7PX5
 
 | | ATAD2 in complex with 1-Methyl-2-quinolone | | Descriptor: | 1,2-ETHANEDIOL, 1-METHYLQUINOLIN-2(1H)-ONE, ATPase family AAA domain-containing protein 2, ... | | Authors: | Martin, M.P, Noble, M.E.N. | | Deposit date: | 2021-10-07 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Exiting the tunnel of uncertainty: crystal soak to validated hit.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1F8W
 
 | |
5XJO
 
 | | Plant receptor ERL1-TMM in complex with peptide EPF1 | | Descriptor: | LRR receptor-like serine/threonine-protein kinase ERL1, Protein EPIDERMAL PATTERNING FACTOR 1, Protein TOO MANY MOUTHS | | Authors: | Chai, J, Lin, G, Zhang, L, Han, Z, Shpak, E.D, Yang, X. | | Deposit date: | 2017-05-03 | | Release date: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2.626 Å) | | Cite: | A receptor-like protein acts as a specificity switch for the regulation of stomatal development.
Genes Dev., 31, 2017
|
|
6RIR
 
 | | Crystal structure of phosphorylated Rab8a in complex with the Rab-binding domain of RILPL2 | | Descriptor: | GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Waschbusch, D, Purlyte, E, Alessi, D.R, Khan, A.R. | | Deposit date: | 2019-04-25 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Structural Basis for Rab8a Recruitment of RILPL2 via LRRK2 Phosphorylation of Switch 2.
Structure, 28, 2020
|
|
1TF3
 
 | | TFIIIA FINGER 1-3 BOUND TO DNA, NMR, 22 STRUCTURES | | Descriptor: | 5S RNA GENE, TRANSCRIPTION FACTOR IIIA, ZINC ION | | Authors: | Foster, M.P, Wuttke, D.S, Radhakrishnan, I, Case, D.A, Gottesfeld, J.M, Wright, P.E. | | Deposit date: | 1997-07-01 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Domain packing and dynamics in the DNA complex of the N-terminal zinc fingers of TFIIIA.
Nat.Struct.Biol., 4, 1997
|
|
6PE4
 
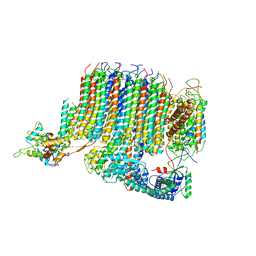 | | Yeast Vo motor in complex with 1 VopQ molecule | | Descriptor: | Cation transporter, Uncharacterized protein YPR170W-B, V-type proton ATPase subunit a, ... | | Authors: | Peng, W, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2019-06-20 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A distinct inhibitory mechanism of the V-ATPase by Vibrio VopQ revealed by cryo-EM.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6X6V
 
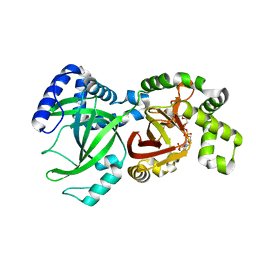 | |
6OZ6
 
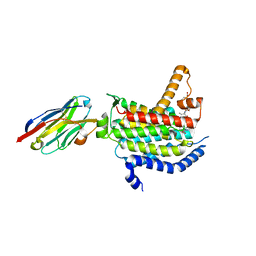 | | Crystal structure of MraY bound to 3'-hydroxymureidomycin A | | Descriptor: | (2~{S})-2-[[(2~{S})-1-[[(2~{S},3~{S})-3-[[(2~{S})-2-azanyl-3-(3-hydroxyphenyl)propanoyl]-methyl-amino]-1-[[(~{Z})-[(3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-ylidene]methyl]amino]-1-oxidanylidene-butan-2-yl]amino]-4-methylsulfanyl-1-oxidanylidene-butan-2-yl]carbamoylamino]-3-(3-hydroxyphenyl)propanoic acid, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2019-05-15 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Chemical logic of MraY inhibition by antibacterial nucleoside natural products.
Nat Commun, 10, 2019
|
|
6XS9
 
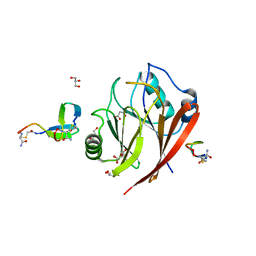 | | Crystal structure of human Vps29 complexed with RaPID-derived cyclic peptide RT-L1 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 48V-TYR-ILE-LYS-THR-PRO-LEU-GLY-THR-PHE-PRO-ASN-ARG-HIS-GLY, GLYCEROL, ... | | Authors: | Chen, K.-E, Guo, Q, Collins, B.M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | De novo macrocyclic peptides for inhibiting, stabilizing, and probing the function of the retromer endosomal trafficking complex.
Sci Adv, 7, 2021
|
|
6PFZ
 
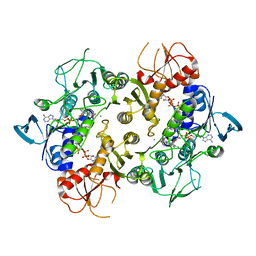 | | Structure of a NAD-Dependent Persulfide Reductase from A. fulgidus | | Descriptor: | CALCIUM ION, COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Sazinsky, M.H, Shabdar, S, Garcia-Constineiras, A, Crane III, E.J. | | Deposit date: | 2019-06-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.10003877 Å) | | Cite: | Structural and Kinetic Characterization of Hyperthermophilic NADH-Dependent Persulfide Reductase from Archaeoglobus fulgidus .
Archaea, 2021, 2021
|
|
8THC
 
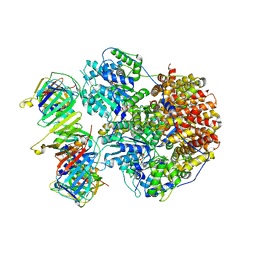 | | Structure of the Saccharomyces cerevisiae clamp unloader Elg1-RFC bound to a cracked PCNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ELG1 isoform 1, MAGNESIUM ION, ... | | Authors: | Zheng, F, Yao, Y.N, Georgescu, R, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-07-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structure of the PCNA unloader Elg1-RFC.
Sci Adv, 10, 2024
|
|
8TYN
 
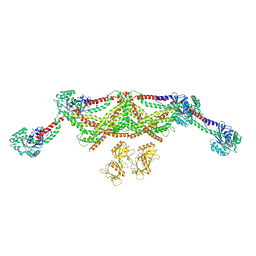 | |
2IT0
 
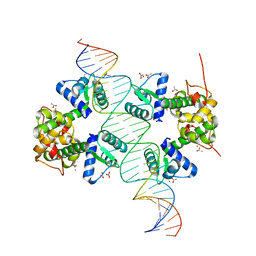 | | Crystal structure of a two-domain IdeR-DNA complex crystal form II | | Descriptor: | ACETATE ION, Iron-dependent repressor ideR, NICKEL (II) ION, ... | | Authors: | Wisedchaisri, G, Chou, C.J, Wu, M, Roach, C, Rice, A.E, Holmes, R.K, Beeson, C, Hol, W.G. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
8TYM
 
 | |
