7A8T
 
 | | Crystal structure of sarcomeric protein FATZ-1 (mini-FATZ-1 construct) in complex with rod domain of alpha-actinin-2 | | Descriptor: | Alpha-actinin-2, Myozenin-1 | | Authors: | Sponga, A, Arolas, J.L, Rodriguez Chamorro, A, Mlynek, G, Hollerl, E, Schreiner, C, Pedron, M, Kostan, J, Ribeiro, E.A, Djinovic-Carugo, K. | | Deposit date: | 2020-08-31 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with alpha-actinin.
Sci Adv, 7, 2021
|
|
6XJ0
 
 | | Crystal structure of multi-copper oxidase from Pediococcus pentosaceus | | Descriptor: | CHLORIDE ION, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Pardo, I, Soares, A.S, Collins, R, Partowmah, S.H, Coler, E.A. | | Deposit date: | 2020-06-22 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural analysis and biochemical properties of laccase enzymes from two Pediococcus species.
Microb Biotechnol, 14, 2021
|
|
5IRV
 
 | | Human cytochrome P450 17A1 bound to inhibitor VT-464 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase, VT-464 | | Authors: | Scott, E.E, Petrunak, E.M. | | Deposit date: | 2016-03-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Structural and Functional Evaluation of Clinically Relevant Inhibitors of Steroidogenic Cytochrome P450 17A1.
Drug Metab. Dispos., 45, 2017
|
|
8ONM
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with D-glutamate | | Descriptor: | (~{Z})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pent-2-enedioic acid, 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing of the structural and catalytic roles of the residues in the active site of transaminase from Aminobacterium colombiense
To Be Published
|
|
6ZOM
 
 | | Oxidized thioredoxin 1 from the anaerobic bacteria Desulfovibrio vulgaris Hildenborough | | Descriptor: | Thioredoxin | | Authors: | Garcin, E, Bornet, O, Nouailler, M, Pieulle, L, Guerlesquin, F, Sebban-Kreuzer, C. | | Deposit date: | 2020-07-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Glutamate optimizes enzymatic activity under high hydrostatic pressure in Desulfovibrio species: effects on the ubiquitous thioredoxin system.
Extremophiles, 25, 2021
|
|
1F5P
 
 | |
6EN3
 
 | | Crystal structure of full length EndoS from Streptococcus pyogenes in complex with G2 oligosaccharide. | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F2,Multifunctional-autoprocessing repeats-in-toxin, NICKEL (II) ION, ... | | Authors: | Trastoy, B, Klontz, E.H, Orwenyo, J, Marina, A, Wang, L.X, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2017-10-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structural basis for the recognition of complex-type N-glycans by Endoglycosidase S.
Nat Commun, 9, 2018
|
|
7A7G
 
 | | Soluble epoxide hydrolase in complex with TK90 | | Descriptor: | (2~{R})-2-[[4-[[4-methoxy-2-(trifluoromethyl)phenyl]methylcarbamoyl]phenyl]methyl]butanoic acid, Bifunctional epoxide hydrolase 2 | | Authors: | Ni, X, Kramer, J.S, Kirchner, T, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Combined Cardioprotective and Adipocyte Browning Effects Promoted by the Eutomer of Dual sEH/PPAR gamma Modulator.
J.Med.Chem., 64, 2021
|
|
7PEG
 
 | | Structure of the sporulation/germination protein YhcN from Bacillus subtilis | | Descriptor: | Probable spore germination lipoprotein YhcN | | Authors: | Liu, B, Chan, H, Bauda, E, Contreras-Martel, C, Bellard, L, Villard, A.M, Mas, C, Neumann, E, Fenel, D, Favier, A, Serrano, M, Henriques, A.O.H, Rodrigues, C.D.A, Morlot, C. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural insights into ring-building motif domains involved in bacterial sporulation.
J.Struct.Biol., 214, 2022
|
|
5I7Y
 
 | | BRD9 in complex with Cpd4 ((E)-3-(6-(but-2-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)-N,N-dimethylbenzamide) | | Descriptor: | 3-{6-[(2E)-but-2-en-1-yl]-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl}-N,N-dimethylbenzamide, Bromodomain-containing protein 9 | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4514 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
7N6B
 
 | | Structure of MmpL3 reconstituted into lipid nanodisc in the TMM bound state | | Descriptor: | 6-O-[(2S)-2-{(1S)-18-[(1R,2R)-2-hexylcyclopropyl]-1-hydroxyoctadecyl}tricosanoyl]-alpha-D-glucopyranosyl alpha-D-glucopyranoside, MmpL3 transporter | | Authors: | Su, C.C, Yu, E. | | Deposit date: | 2021-06-08 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structures of the mycobacterial membrane protein MmpL3 reveal its mechanism of lipid transport.
Plos Biol., 19, 2021
|
|
1FFO
 
 | | CRYSTAL STRUCTURE OF MURINE CLASS I H-2DB COMPLEXED WITH SYNTHETIC PEPTIDE GP33 (C9M/K1A) | | Descriptor: | BETA-2 MICROGLOBULIN BETA CHAIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B, ... | | Authors: | Wang, B, Sharma, A, Maile, R, Saad, M, Collins, E.J, Frelinger, J.A. | | Deposit date: | 2000-07-25 | | Release date: | 2002-12-11 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Peptidic termini play a significant role in TCR recognition.
J.Immunol., 169, 2002
|
|
1F6S
 
 | | CRYSTAL STRUCTURE OF BOVINE ALPHA-LACTALBUMIN | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION | | Authors: | Chrysina, E.D, Brew, K, Acharya, K.R. | | Deposit date: | 2000-06-23 | | Release date: | 2000-12-13 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of apo- and holo-bovine alpha-lactalbumin at 2. 2-A resolution reveal an effect of calcium on inter-lobe interactions.
J.Biol.Chem., 275, 2000
|
|
7PTQ
 
 | | RNA origami 5-helix tile | | Descriptor: | Chains: C | | Authors: | McRae, E.K.S, Andersen, E.S. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structure, folding and flexibility of co-transcriptional RNA origami.
Nat Nanotechnol, 18, 2023
|
|
8TL6
 
 | | Cryo-EM structure of DDB1deltaB-DDA1-DCAF5 | | Descriptor: | DDB1- and CUL4-associated factor 5, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1 | | Authors: | Yue, H, Hunkeler, M, Roy Burman, S.S, Fischer, E.S. | | Deposit date: | 2023-07-26 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Targeting DCAF5 suppresses SMARCB1-mutant cancer by stabilizing SWI/SNF.
Nature, 628, 2024
|
|
6UAX
 
 | | Crystal structure of a GH128 (subgroup II) endo-beta-1,3-glucanase from Sorangium cellulosum (ScGH128_II) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Santos, C.R, Costa, P.A.C.R, Domingues, M.N, Lima, E.A, Mandelli, F, Vieira, P.S, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UB6
 
 | | Crystal structure of a GH128 (subgroup IV) endo-beta-1,3-glucanase from Lentinula edodes (LeGH128_IV) in complex with laminaritetraose | | Descriptor: | CHLORIDE ION, Endo-beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Santos, C.R, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2020-08-05 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
4XR7
 
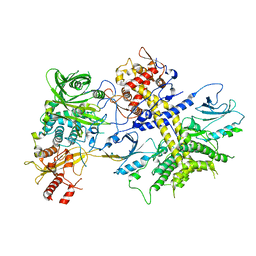 | | Structure of the Saccharomyces cerevisiae PAN2-PAN3 core complex | | Descriptor: | PAB-dependent poly(A)-specific ribonuclease subunit PAN2, PAB-dependent poly(A)-specific ribonuclease subunit PAN3 | | Authors: | Schafer, I.B, Rode, M, Bonneau, F, Schussler, S, Conti, E. | | Deposit date: | 2015-01-20 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.796 Å) | | Cite: | The structure of the Pan2-Pan3 core complex reveals cross-talk between deadenylase and pseudokinase.
Nat. Struct. Mol. Biol., 21, 2014
|
|
6RSY
 
 | |
6PJX
 
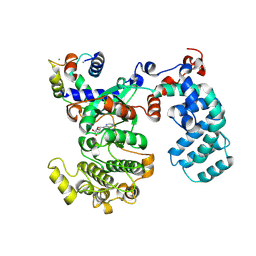 | | Crystal Structure of G Protein-Coupled Receptor Kinase 5 (GRK5) in Complex with Calmodulin (CaM) | | Descriptor: | CALCIUM ION, Calmodulin, G protein-coupled receptor kinase 5, ... | | Authors: | Bhardwaj, A, Komolov, K.E, Sulon, S, Benovic, J.L. | | Deposit date: | 2019-06-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of a GRK5-Calmodulin Complex Reveals Molecular Mechanism of GRK Activation and Substrate Targeting.
Mol.Cell, 81, 2021
|
|
5TXL
 
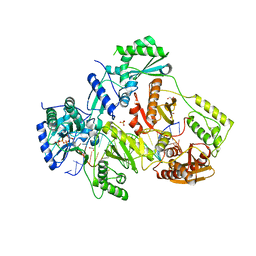 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DATP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG) P*CP*GP*CP*CP*GP)-3'), ... | | Authors: | Das, K, Martinez, S.M, Arnold, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
5MBQ
 
 | | CeuE (H227A variant) a periplasmic protein from Campylobacter jejuni | | Descriptor: | Enterochelin uptake periplasmic binding protein | | Authors: | Wilde, E.J, Blagova, E.V, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
7PTS
 
 | | RNA origami 5-helix tile | | Descriptor: | 5HT-B | | Authors: | McRae, E.K.S, Andersen, E.S. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.71 Å) | | Cite: | Structure, folding and flexibility of co-transcriptional RNA origami.
Nat Nanotechnol, 18, 2023
|
|
6ENM
 
 | | Crystal structure of MMP12 in complex with hydroxamate inhibitor LP168. | | Descriptor: | 2-[2-[4-(4-methoxyphenyl)phenyl]sulfonylphenyl]-~{N}-oxidanyl-ethanamide, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Vera, L, Nuti, E, Rossello, A, Stura, E.A. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of Thioaryl-Based Matrix Metalloproteinase-12 Inhibitors with Alternative Zinc-Binding Groups: Synthesis, Potentiometric, NMR, and Crystallographic Studies.
J. Med. Chem., 61, 2018
|
|
7S7A
 
 | | Crystal structure of CDK2 liganded with compound EF3019 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(2H-indazol-3-yl)ethyl]amino}-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
