8UMT
 
 | | Atomic model of the human CTF18-RFC-PCNA binary complex in the three-subunit binding state (state 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, MAGNESIUM ION, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UMY
 
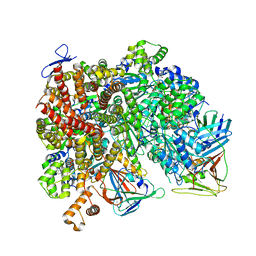 | | Atomic model of the human CTF18-RFC-PCNA-DNA ternary complex with narrow PCNA opening state II (state 6) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, DNA (20-MER), ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UN0
 
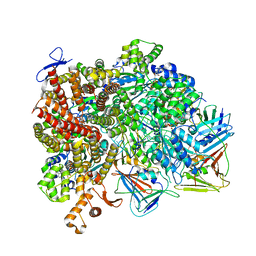 | | Atomic model of the human CTF18-RFC-PCNA-DNA ternary complex with cracked and closed PCNA (state 7) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, DNA (20-MER), ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7MHT
 
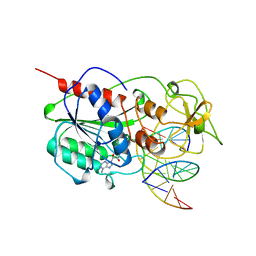 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | Descriptor: | 5'-D(P*CP*CP*AP*TP*GP*AP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | Authors: | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | Deposit date: | 1998-08-05 | | Release date: | 1998-11-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
4HGK
 
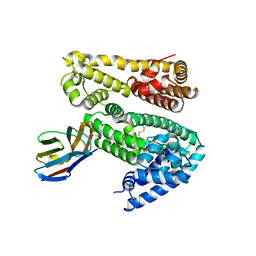 | | Shark IgNAR variable domain | | Descriptor: | Serum albumin, shark V-NAR antibody | | Authors: | Olland, A.O, Kovalenko, O.V, Svenson, K, King, D. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Atypical Antigen Recognition Mode of a Shark Immunoglobulin New Antigen Receptor (IgNAR) Variable Domain Characterized by Humanization and Structural Analysis.
J.Biol.Chem., 288, 2013
|
|
7MHL
 
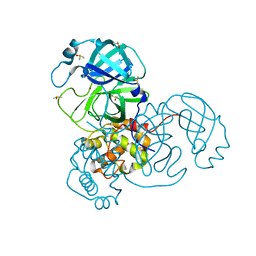 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 100 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7SHW
 
 | |
7SF8
 
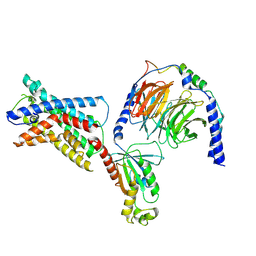 | | GPR56 (ADGRG1) 7TM domain bound to tethered agonist in complex with G protein heterotrimer | | Descriptor: | G protein subunit 13 (Gi2-mini-G13 chimera), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Barros-Alvarez, X, Panova, O, Skiniotis, G. | | Deposit date: | 2021-10-03 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The tethered peptide activation mechanism of adhesion GPCRs.
Nature, 604, 2022
|
|
7SF7
 
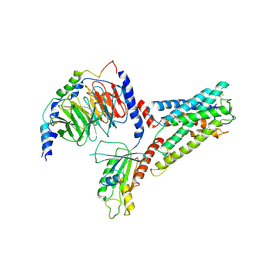 | | LPHN3 (ADGRL3) 7TM domain bound to tethered agonist in complex with G protein heterotrimer | | Descriptor: | G protein subunit 13 (Gi2-mini-G13 chimera), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Barros-Alvarez, X, Panova, O, Skiniotis, G. | | Deposit date: | 2021-10-03 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The tethered peptide activation mechanism of adhesion GPCRs.
Nature, 604, 2022
|
|
4HGD
 
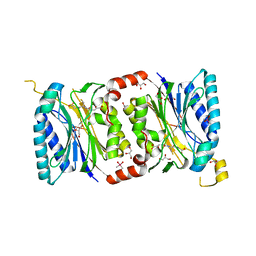 | | Structural insights into yeast Nit2: C169S mutant of yeast Nit2 in complex with an endogenous peptide-like ligand | | Descriptor: | CACODYLATE ION, GLYCEROL, N-(4-carboxy-4-oxobutanoyl)-L-cysteinylglycine, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-08 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8UI8
 
 | |
7MKF
 
 | |
4K2C
 
 | | HSA Ligand Free | | Descriptor: | Serum albumin | | Authors: | Wang, Y, Luo, Z, Shi, X, Huang, M. | | Deposit date: | 2013-04-08 | | Release date: | 2013-05-01 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural mechanism of ring-opening reaction of glucose by human serum albumin.
J. Biol. Chem., 288, 2013
|
|
7MKH
 
 | |
7MKG
 
 | |
4HGM
 
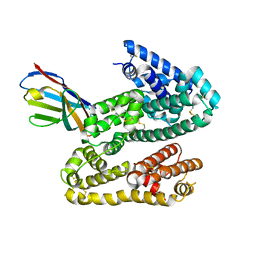 | | Shark IgNAR Variable Domain | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, Serum albumin, ... | | Authors: | Olland, A, Kovalenko, O.V, King, D, Svenson, K. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Atypical Antigen Recognition Mode of a Shark Immunoglobulin New Antigen Receptor (IgNAR) Variable Domain Characterized by Humanization and Structural Analysis.
J.Biol.Chem., 288, 2013
|
|
5Z37
 
 | |
4HG3
 
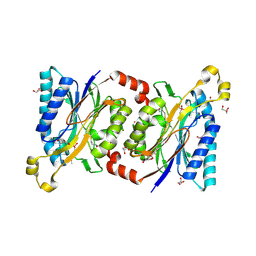 | | Structural insights into yeast Nit2: wild-type yeast Nit2 in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CACODYLATE ION, GLYCEROL, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-06 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4K2G
 
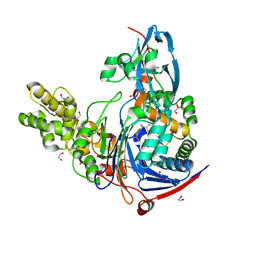 | | Structure of Pseudomonas aeruginosa PvdQ bound to BRD-A33442372 | | Descriptor: | (2S)-(4-fluorophenyl)[6-(trifluoromethyl)pyridin-2-yl]ethanenitrile, 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase PvdQ | | Authors: | Drake, E.J, Wurst, J.M, Theriault, J.R, Munoz, B, Gulick, A.M. | | Deposit date: | 2013-04-09 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Inhibitors of PvdQ, an Enzyme Involved in the Synthesis of the Siderophore Pyoverdine.
Acs Chem.Biol., 9, 2014
|
|
7LZ0
 
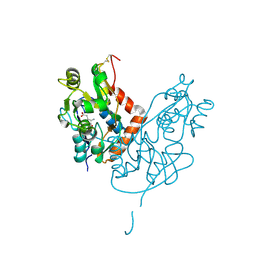 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
8UAK
 
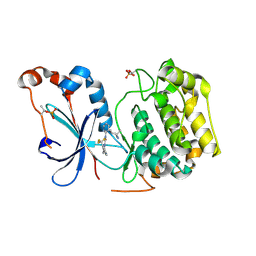 | | Crystal structure of the catalytic domain of human PKC alpha (D463N, V568I, S657E) in complex with Darovasertib (NVP-LXS196) at 2.82-A resolution | | Descriptor: | (6M)-3-amino-N-[3-(4-amino-4-methylpiperidin-1-yl)pyridin-2-yl]-6-[3-(trifluoromethyl)pyridin-2-yl]pyrazine-2-carboxamide, Protein kinase C alpha type | | Authors: | Romanowski, M.J, Lam, J, Visser, M. | | Deposit date: | 2023-09-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of Darovasertib (NVP-LXS196), a Pan-PKC Inhibitor for the Treatment of Metastatic Uveal Melanoma.
J.Med.Chem., 67, 2024
|
|
7KWW
 
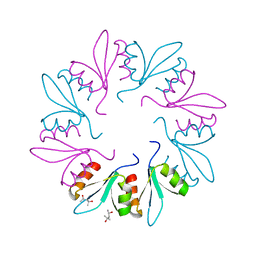 | | X-ray Crystal Structure of PlyCB Mutant K59H | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7KWY
 
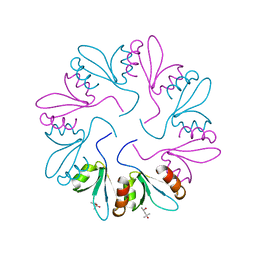 | | X-ray Crystal Structure of PlyCB Mutant R66K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7KWT
 
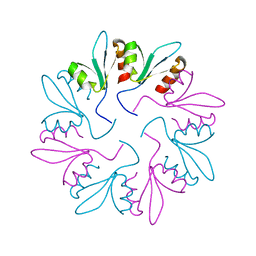 | | X-ray Crystal Structure of PlyCB Mutant Y28H | | Descriptor: | PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
8WK0
 
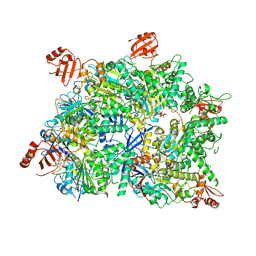 | | Cryo-EM structure of a bacterial protein | | Descriptor: | Helicase HerA central domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Yu, G, Liao, F, Li, X, Li, Z, Zhang, H. | | Deposit date: | 2023-09-26 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of a bacterial protein
To Be Published
|
|
