5G1C
 
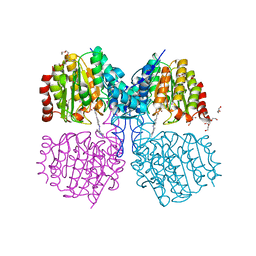 | | Structure of HDAC like protein from Bordetella Alcaligenes bound the photoswitchable pyrazole Inhibitor CEW395 | | Descriptor: | (2E)-N-hydroxy-3-{4-[(E)-(1,3,5-trimethyl-1H-pyrazol-4-yl)diazenyl]phenyl}prop-2-enamide, DI(HYDROXYETHYL)ETHER, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-24 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Toward Photopharmacological Antimicrobial Chemotherapy Using Photoswitchable Amidohydrolase Inhibitors.
ACS Infect Dis, 3, 2017
|
|
9CCS
 
 | |
9CCW
 
 | |
9CCU
 
 | |
9CCT
 
 | |
5FKA
 
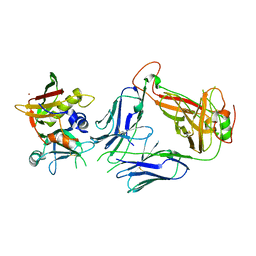 | | Crystal structure of staphylococcal enterotoxin E in complex with a T cell receptor | | Descriptor: | STAPHYLOCOCCAL ENTEROTOXIN E, T CELL RECEPTOR ALPHA CHAIN, T CELL RECEPTOR BETA CHAIN, ... | | Authors: | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | Deposit date: | 2015-10-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two Common Structural Motifs for Tcr Recognition by Staphylococcal Enterotoxins.
Sci.Rep., 6, 2016
|
|
6L0R
 
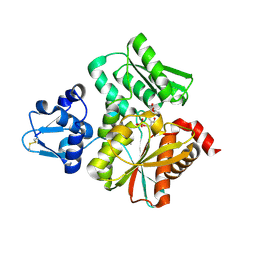 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Acetylserine | | Descriptor: | (2S)-3-acetyloxy-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
5FKZ
 
 | | Structure of E.coli Constitutive lysine decarboxylase | | Descriptor: | LYSINE DECARBOXYLASE, CONSTITUTIVE | | Authors: | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | Deposit date: | 2015-10-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
5FMG
 
 | | Structure and function based design of Plasmodium-selective proteasome inhibitors | | Descriptor: | (2S)-N-[(E,2S)-1-(1H-indol-3-yl)-4-methylsulfonyl-but-3-en-2-yl]-2-[[(2S)-3-(1H-indol-3-yl)-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]-4-methyl-pentanamide, BETA3 PROTEASOME SUBUNIT, PUTATIVE, ... | | Authors: | Li, H, O'Donoghue, A.J, van der Linden, W.A, Xie, S.C, Yoo, E, Foe, I.T, Tilley, L, Craik, C.S, da Fonseca, P.C.A, Bogyo, M. | | Deposit date: | 2015-11-04 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and Function Based Design of Plasmodium-Selective Proteasome Inhibitors
Nature, 530, 2016
|
|
1LLR
 
 | | CHOLERA TOXIN B-PENTAMER WITH LIGAND BMSC-0012 | | Descriptor: | 3-AMINO-4-{3-[2-(2-PROPOXY-ETHOXY)-ETHOXY]-PROPYLAMINO}-CYCLOBUT-3-ENE-1,2-DIONE, 5-aminocarbonyl-3-nitrophenyl alpha-D-galactopyranoside, CHOLERA TOXIN B SUBUNIT | | Authors: | Merritt, E.A, Hol, W.G.J. | | Deposit date: | 2002-04-30 | | Release date: | 2002-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Characterization and crystal structure of a high-affinity pentavalent receptor-binding inhibitor for cholera toxin and E. coli heat-labile enterotoxin.
J.Am.Chem.Soc., 124, 2002
|
|
1T5C
 
 | | Crystal structure of the motor domain of human kinetochore protein CENP-E | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Centromeric protein E, MAGNESIUM ION, ... | | Authors: | Garcia-Saez, I, Yen, T, Wade, R.H, Kozielski, F, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-05-04 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the motor domain of the human kinetochore protein CENP-E.
J.Mol.Biol., 340, 2004
|
|
6APV
 
 | |
5J18
 
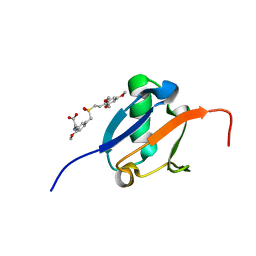 | | Solution structure of Ras Binding Domain (RBD) of B-Raf complexed with Rigosertib (Complex I) | | Descriptor: | N-[2-methoxy-5-({[(E)-2-(2,4,6-trimethoxyphenyl)ethenyl]sulfonyl}methyl)phenyl]glycine, Serine/threonine-protein kinase B-raf | | Authors: | Dutta, K, Vasquez-Del Carpio, R, Aggarwal, A.K, Reddy, E.P. | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule RAS-Mimetic Disrupts RAS Association with Effector Proteins to Block Signaling.
Cell, 165, 2016
|
|
7LXJ
 
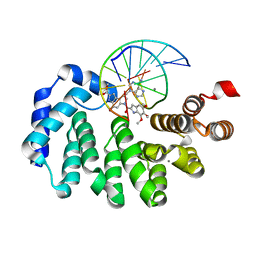 | |
6L0S
 
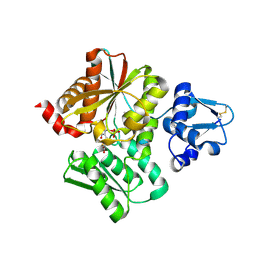 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with L-Cysteine | | Descriptor: | (2R)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-sulfanyl-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
2JX2
 
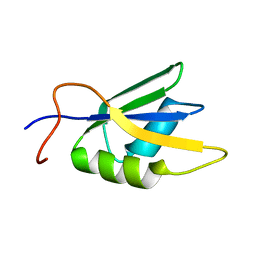 | | Solution conformation of RNA-bound NELF-E RRM | | Descriptor: | Negative elongation factor E | | Authors: | Jampani, N, Schweimer, K, Wenzel, S, Woehrl, B.M, Roesch, P. | | Deposit date: | 2007-11-02 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NELF-E RRM Undergoes Major Structural Changes in Flexible Protein Regions on Target RNA Binding
Biochemistry, 47, 2008
|
|
7N6K
 
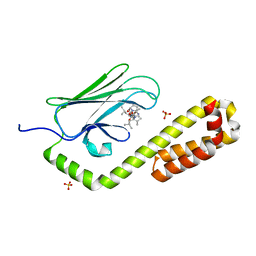 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RALALLPLSR | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2021-06-08 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6L0Q
 
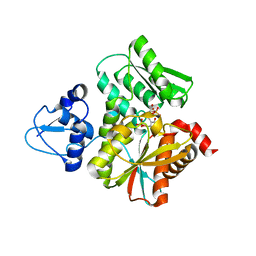 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Phosphoserine | | Descriptor: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
7N6J
 
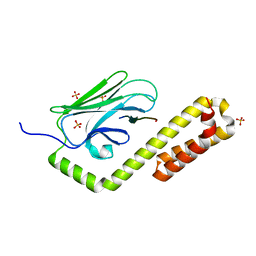 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RKQSTIALALLPLLFTPRR | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2021-06-08 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N6L
 
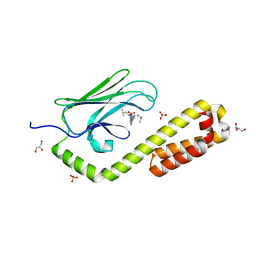 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide EANQQKPLLGLFADG | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, GLYCEROL, ... | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2021-06-08 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6F6U
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-7b | | Descriptor: | 2-[(~{E})-(3-cyclopentyloxy-4-methoxy-phenyl)methylideneamino]oxy-1-[(2~{R},6~{S})-2,6-dimethylmorpholin-4-yl]ethanone, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6L0P
 
 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Phosphoserine | | Descriptor: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
2JAM
 
 | | Crystal structure of human calmodulin-dependent protein kinase I G | | Descriptor: | 1,2-ETHANEDIOL, 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, CALCIUM ION, ... | | Authors: | Debreczeni, J.E, Bullock, A, Keates, T, Niesen, F.H, Salah, E, Shrestha, L, Smee, C, Sobott, F, Pike, A.C.W, Bunkoczi, G, von Delft, F, Turnbull, A, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Knapp, S. | | Deposit date: | 2006-11-29 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Calmodulin-Dependent Protein Kinase I G
To be Published
|
|
4PLK
 
 | |
6F8T
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-4a | | Descriptor: | (2~{R})-1-[(~{E})-(3-cyclopentyloxy-4-methoxy-phenyl)methylideneamino]oxy-3-[(2~{R},6~{S})-2,6-dimethylmorpholin-4-yl]propan-2-ol, MAGNESIUM ION, ZINC ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
