8BJH
 
 | | chimera of the inactive ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, with the double mutation K3528M and K3535I, fused to a proline-Rich-Domain (PRD) and profilin, bound to Latrunculin B-ADP-Mg-actin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
8BO1
 
 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to Latrunculin-B-ATP-Mg-actin, and 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE and 2 Mg ions | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AZIDE ION, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
8BJI
 
 | | chimera of ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo fused to a proline-Rich-Domain (PRD) and profilin, bound to ADP-Mg-actin and a sulfate ion | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
8BR1
 
 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to Latrunculin-B-ATP-Mg-actin, and 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE and 2 Mg ions | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-22 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
8BJJ
 
 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin, bound to ATP-Mg-actin, human profilin 1 and a sulfate ion | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Teixeira-Nunes, M, Renault, L, Retailleau, P. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
8BR0
 
 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin (residue Q3455 to L3863) in complex with 3'deoxyCTP and two manganese cations bound to Latrunculin-B-ADP-Mn-actin | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Texeira-Nuns, M, Retailleau, P, Renault, L. | | Deposit date: | 2022-11-22 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
2WWM
 
 | | Crystal structure of the Titin M10-Obscurin like 1 Ig complex in space group P1 | | Descriptor: | OBSCURIN-LIKE PROTEIN 1, TITIN | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insight Into M-Band Assembly and Mechanics from the Titin-Obscurin-Like-1 Complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WP3
 
 | | Crystal structure of the Titin M10-Obscurin like 1 Ig complex | | Descriptor: | GLYCEROL, OBSCURIN-LIKE PROTEIN 1, SULFATE ION, ... | | Authors: | Pernigo, S, Fuzukawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2009-08-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Insight Into M-Band Assembly and Mechanics from the Titin-Obscurin-Like-1 Complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6TZ8
 
 | |
6TZ6
 
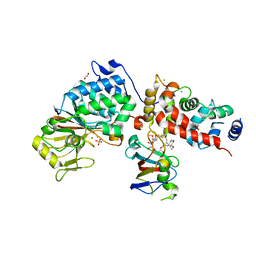 | | Crystal Structure of Candida Albicans Calcineurin A, Calcineurin B, FKBP12 and FK506 (Tacrolimus) | | Descriptor: | 1,2-ETHANEDIOL, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, ... | | Authors: | Fox III, D, Lukacs, C.M. | | Deposit date: | 2019-08-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents.
Nat Commun, 10, 2019
|
|
1UUE
 
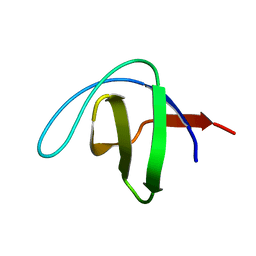 | | a-SPECTRIN SH3 DOMAIN (V44T, D48G MUTANT) | | Descriptor: | SPECTRIN ALPHA CHAIN | | Authors: | Vega, M.C, Fernandez, A, Wilmanns, M, Serrano, L. | | Deposit date: | 2003-12-18 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Solvation in Protein Folding Analysis: Combination of Theoretical and Experimental Approaches
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1WGP
 
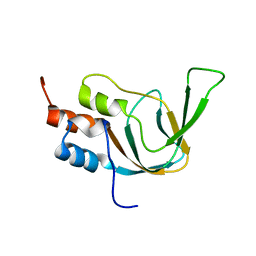 | | Solution structure of the cNMP-binding domain from Arabidopsis thaliana cyclic nucleotide-regulated ion channel | | Descriptor: | Probable cyclic nucleotide-gated ion channel 6 | | Authors: | Chikayama, E, Nameki, N, Kigawa, T, Koshiba, S, Inoue, M, Tomizawa, T, Kobayashi, N, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cNMP-binding domain from Arabidopsis thaliana cyclic nucleotide-regulated ion channel
To be Published
|
|
1WKU
 
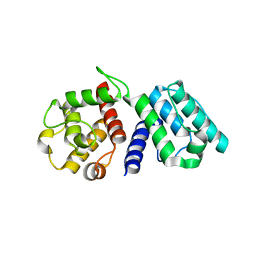 | |
2M1U
 
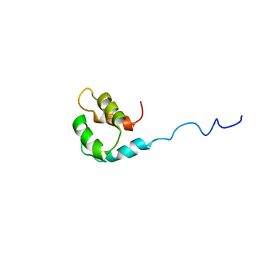 | | Solution structure of the small dictyostelium discoideium myosin light chain mlcb provides insights into iq-motif recognition of class i myosin myo1b | | Descriptor: | myosin light chain MlcB | | Authors: | Liburd, J, Chitayat, S, Crawley, S.W, Denis, C.M, Cote, G.P, Smith, S.P. | | Deposit date: | 2012-12-06 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the small Dictyostelium discoideum myosin light chain MlcB provides insights into MyoB IQ motif recognition.
J.Biol.Chem., 289, 2014
|
|
2M8U
 
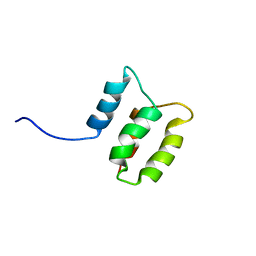 | | Solution structure of the Dictyostelium discodieum Myosin Light Chain, MlcC | | Descriptor: | Myosin Light Chain, MlcC | | Authors: | Liburd, J.D, Miller, E, Langelaan, D, Chitayat, S, Crawley, S.W, Cote, G.P, Smith, S.P. | | Deposit date: | 2013-05-28 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Single-lobe Myosin Light Chain C in Complex with the Light Chain-binding Domains of Myosin-1C Provides Insights into Divergent IQ Motif Recognition.
J.Biol.Chem., 291, 2016
|
|
1WY9
 
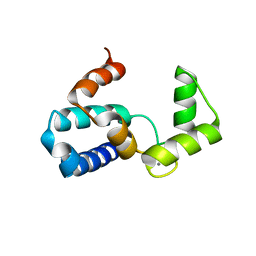 | | Crystal structure of microglia-specific protein, Iba1 | | Descriptor: | Allograft inflammatory factor 1, CALCIUM ION | | Authors: | Yamada, M, Imai, Y, Kohsaka, S, Kamitori, S. | | Deposit date: | 2005-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structures of the Microglia/Macrophage-specific Protein Iba1 from Human and Mouse Demonstrate Novel Molecular Conformation Change Induced by Calcium binding
J.Mol.Biol., 364, 2006
|
|
1DGU
 
 | |
1W2F
 
 | | Human Inositol (1,4,5)-trisphosphate 3-kinase substituted with selenomethionine | | Descriptor: | INOSITOL-TRISPHOSPHATE 3-KINASE A, SULFATE ION | | Authors: | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
1WAA
 
 | | IG27 protein domain | | Descriptor: | TITIN, ZINC ION | | Authors: | Vega, M.C, Valencia, L, Zou, P, Wilmanns, M. | | Deposit date: | 2004-10-25 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanical Network in Titin Immunoglobulin from Force Distribution Analysis.
Plos Comput.Biol., 5, 2009
|
|
1W7J
 
 | | Crystal Structure Of Myosin V Motor With Essential Light Chain + ADP-BeFx - Near Rigor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Coureux, P.-D, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2004-09-03 | | Release date: | 2005-02-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three Myosin V Structures Delineate Essential Features of Chemo-Mechanical Transduction
Embo J., 23, 2004
|
|
1DGV
 
 | |
4RAG
 
 | | CRYSTAL STRUCTURE of PPC2A-D38K | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 1A | | Authors: | Pan, C, Tang, J.Y, Xu, Y.F, Xiao, P, Liu, H.D, Wang, H.A, Wang, W.B, Meng, F.G, Yu, X, Sun, J.P. | | Deposit date: | 2014-09-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The catalytic role of the M2 metal ion in PP2C alpha
Sci Rep, 5, 2015
|
|
2ONL
 
 | | Crystal Structure of the p38a-MAPKAP kinase 2 Heterodimer | | Descriptor: | MAP kinase-activated protein kinase 2, Mitogen-activated protein kinase 14 | | Authors: | Ter Haar, E. | | Deposit date: | 2007-01-24 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of the P38alpha-MAPKAP kinase 2 heterodimer.
J.Biol.Chem., 282, 2007
|
|
4JS9
 
 | | Structural Characterization of Inducible Nitric Oxide Synthase Substituted With Mesoheme | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, Mesoheme, Nitric oxide synthase, ... | | Authors: | Hannibal, L, Page, R.C, Bolisetty, K, Yu, Z, Misra, S, Stuehr, D.J. | | Deposit date: | 2013-03-22 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Kinetic and Structural Characterization of Inducible Nitric Oxide Synthase Substituted With Mesoheme
To be Published
|
|
7AW8
 
 | | Alpha-actinin in Rhodamnia argentea | | Descriptor: | ACETIC ACID, Alpha actinin, actin binding domain, ... | | Authors: | Persson, K, Backman, L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and functional characterization of a plant alpha-actinin.
Febs Open Bio, 11, 2021
|
|
