6NWE
 
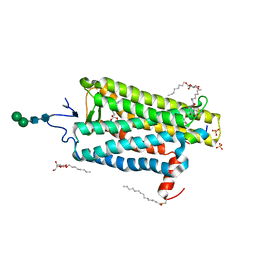 | |
6NI3
 
 | | B2V2R-Gs protein subcomplex of a GPCR-G protein-beta-arrestin mega-complex | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Endolysin,Beta-2 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Nguyen, A.H, Thomsen, A.R.B, Cahill, T.J, des Georges, A, Lefkowitz, R.J. | | Deposit date: | 2018-12-26 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of an endosomal signaling GPCR-G protein-beta-arrestin megacomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6N4B
 
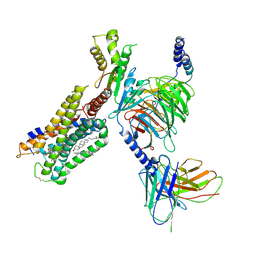 | | Cannabinoid Receptor 1-G Protein Complex | | Descriptor: | CHOLESTEROL, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Krishna Kumar, K, Shalev-Benami, M, Hu, H, Weis, W.I, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2018-11-18 | | Release date: | 2019-01-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a Signaling Cannabinoid Receptor 1-G Protein Complex.
Cell, 176, 2019
|
|
6N48
 
 | | Structure of beta2 adrenergic receptor bound to BI167107, Nanobody 6B9, and a positive allosteric modulator | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid Antibody Fragment, ... | | Authors: | Liu, X, Masoudi, A, Kahsai, A.W, Huang, L.Y, Pani, B, Hirata, K, Ahn, S, Lefkowitz, R.J, Kobilka, B.K. | | Deposit date: | 2018-11-17 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of beta2AR regulation by an intracellular positive allosteric modulator.
Science, 364, 2019
|
|
6MXT
 
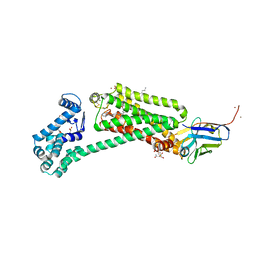 | | Crystal structure of human beta2 adrenergic receptor bound to salmeterol and Nb71 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endolysin, ... | | Authors: | Masureel, M, Zou, Y, Picard, L.P, van der Westhuizen, E, Mahoney, J.P, Rodrigues, J.P.G.L.M, Mildorf, T.J, Dror, R.O, Shaw, D.E, Bouvier, M, Pardon, E, Steyaert, J, Sunahara, R.K, Weis, W.I, Zhang, C, Kobilka, B.K. | | Deposit date: | 2018-10-31 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95934224 Å) | | Cite: | Structural insights into binding specificity, efficacy and bias of a beta2AR partial agonist.
Nat. Chem. Biol., 14, 2018
|
|
6MH8
 
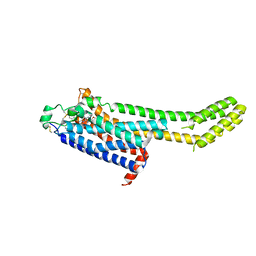 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, Soluble cytochrome b562 chimeric construct | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
6MET
 
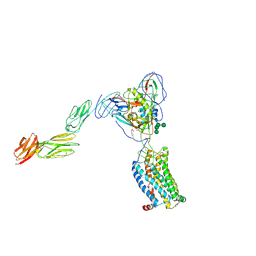 | | Structural basis of coreceptor recognition by HIV-1 envelope spike | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shaik, M.M, Chen, B. | | Deposit date: | 2018-09-07 | | Release date: | 2018-12-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of coreceptor recognition by HIV-1 envelope spike.
Nature, 565, 2018
|
|
6MEO
 
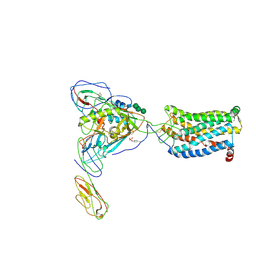 | | Structural basis of coreceptor recognition by HIV-1 envelope spike | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shaik, M.M, Chen, B. | | Deposit date: | 2018-09-06 | | Release date: | 2018-12-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of coreceptor recognition by HIV-1 envelope spike.
Nature, 565, 2018
|
|
6ME9
 
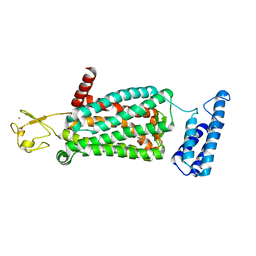 | | XFEL crystal structure of human melatonin receptor MT2 in complex with ramelteon | | Descriptor: | N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}propanamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ZINC ION | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
6ME8
 
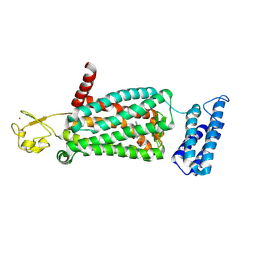 | | XFEL crystal structure of human melatonin receptor MT2 (N86D) in complex with 2-phenylmelatonin | | Descriptor: | N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ZINC ION | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
6ME7
 
 | | XFEL crystal structure of human melatonin receptor MT2 (H208A) in complex with 2-phenylmelatonin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ... | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
6ME6
 
 | | XFEL crystal structure of human melatonin receptor MT2 in complex with 2-phenylmelatonin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ... | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
6ME5
 
 | | XFEL crystal structure of human melatonin receptor MT1 in complex with agomelatine | | Descriptor: | OLEIC ACID, chimera protein of Melatonin receptor type 1A and GlgA glycogen synthase, ~{N}-[2-(7-methoxynaphthalen-1-yl)ethyl]ethanamide | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
6ME4
 
 | | XFEL crystal structure of human melatonin receptor MT1 in complex with 2-iodomelatonin | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-[2-(2-iodo-5-methoxy-1H-indol-3-yl)ethyl]acetamide, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
6ME3
 
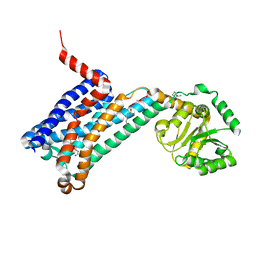 | | XFEL crystal structure of human melatonin receptor MT1 in complex with 2-phenylmelatonin | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, OLEIC ACID, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
6ME2
 
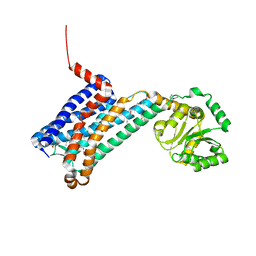 | | XFEL crystal structure of human melatonin receptor MT1 in complex with ramelteon | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}propanamide, OLEIC ACID, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
6M9T
 
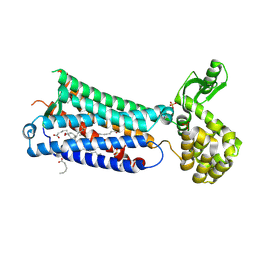 | | Crystal structure of EP3 receptor bound to misoprostol-FA | | Descriptor: | (11alpha,12alpha,13E,16S)-11,16-dihydroxy-16-methyl-9-oxoprost-13-en-1-oic acid, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, OLEIC ACID, ... | | Authors: | Audet, M, White, K.L, Breton, B, Zarzycka, B, Han, G.W, Lu, Y, Gati, C, Batyuk, A, Popov, P, Velasquez, J, Manahan, D, Hu, H, Weierstall, U, Liu, W, Shui, W, Katrich, V, Cherezov, V, Hanson, M.A, Stevens, R.C. | | Deposit date: | 2018-08-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of misoprostol bound to the labor inducer prostaglandin E2receptor.
Nat. Chem. Biol., 15, 2019
|
|
6LW5
 
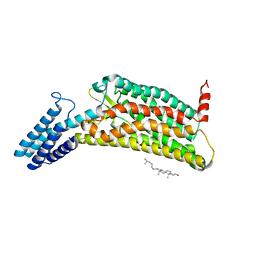 | | Crystal structure of the human formyl peptide receptor 2 in complex with WKYMVm | | Descriptor: | CHOLESTEROL, Soluble cytochrome b562,N-formyl peptide receptor 2, TRP-LYS-TYR-MET-VAL-QXV | | Authors: | Chen, T, Zong, X, Zhang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2020-02-07 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand binding modes at the human formyl peptide receptor 2.
Nat Commun, 11, 2020
|
|
6LUQ
 
 | | Haloperidol bound D2 dopamine receptor structure inspired discovery of subtype selective ligands | | Descriptor: | 4-[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]-1-(4-fluorophenyl)butan-1-one, OLEIC ACID, chimera of D(2) dopamine receptor and Endolysin | | Authors: | Fan, L, Tan, L, Chen, Z, Qi, J, Nie, F, Luo, Z, Cheng, J, Wang, S. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Haloperidol bound D2dopamine receptor structure inspired the discovery of subtype selective ligands.
Nat Commun, 11, 2020
|
|
6LRY
 
 | | Crystal structure of human endothelin ETB receptor in complex with sarafotoxin S6b | | Descriptor: | Endothelin receptor type B,Endolysin,Endothelin receptor type B, Sarafotoxin-B | | Authors: | Izume, T, Miyauchi, H, Shihoya, W, Nureki, O. | | Deposit date: | 2020-01-16 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human endothelin ETBreceptor in complex with sarafotoxin S6b.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
6LPL
 
 | | A2AR crystallized in EROCOC17+4, SS-ROX at 100 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-11 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
6LPK
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 293 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
6LPJ
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 277 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
6LI3
 
 | | cryo-EM structure of GPR52-miniGs-NB35 | | Descriptor: | G-protein coupled receptor 52, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, M, Wang, N, Xu, F, Wu, J, Lei, M. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LI2
 
 | | Crystal structure of GPR52 ligand free form with rubredoxin fusion | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Rubredoxin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
