1UWB
 
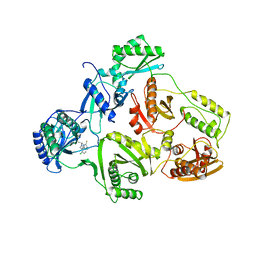 | | TYR 181 CYS HIV-1 RT/8-CL TIBO | | Descriptor: | 5-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Das, K, Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 1996-11-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant.
J.Mol.Biol., 264, 1996
|
|
5TCO
 
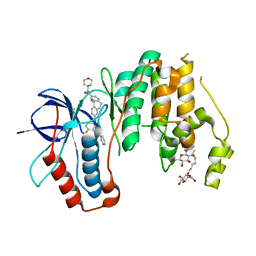 | | Human p38 MAP Kinase in Complex with Dibenzosuberone Compound 1 | | Descriptor: | 3-[(3-benzamido-4-fluoranyl-phenyl)amino]-~{N}-(2-morpholin-4-ylethyl)-11-oxidanylidene-5,6-dihydrodibenzo[1,2-~{d}:1',2'-~{f}][7]annulene-9-carboxamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Mayer-Wrangowski, S.C, Rauh, D. | | Deposit date: | 2016-09-15 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimized Target Residence Time: Type I1/2 Inhibitors for p38 alpha MAP Kinase with Improved Binding Kinetics through Direct Interaction with the R-Spine.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7OBE
 
 | |
3KIK
 
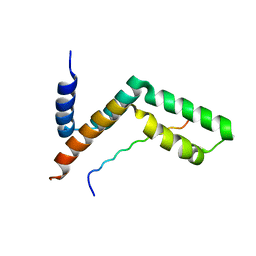 | | Sgf11:Sus1 complex | | Descriptor: | Protein SUS1, SAGA-associated factor 11 | | Authors: | Stewart, M, Ellisdon, A.M. | | Deposit date: | 2009-11-02 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1
J.Biol.Chem., 285, 2010
|
|
5AX3
 
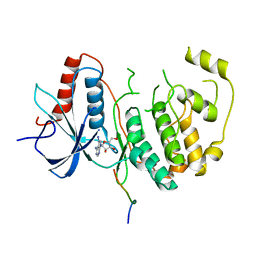 | | Crystal structure of ERK2 complexed with allosteric and ATP-competitive inhibitors. | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Mitogen-activated protein kinase 1, allosteric and ATP-competitive inhibitor | | Authors: | Kinoshita, T, Sugiyama, H, Mori, Y, Takahashi, N, Tomonaga, A. | | Deposit date: | 2015-07-14 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | Identification of allosteric ERK2 inhibitors through in silico biased screening and competitive binding assay
Bioorg.Med.Chem.Lett., 26, 2016
|
|
1A4P
 
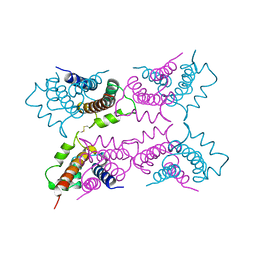 | | P11 (S100A10), LIGAND OF ANNEXIN II | | Descriptor: | S100A10 | | Authors: | Rety, S, Sopkova, J, Renouard, M, Osterloh, D, Gerke, V, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 1998-01-30 | | Release date: | 1998-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of a complex of p11 with the annexin II N-terminal peptide.
Nat.Struct.Biol., 6, 1999
|
|
2GFS
 
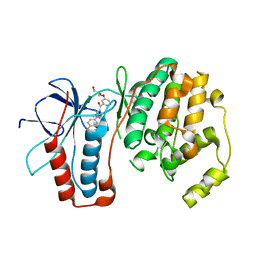 | | P38 Kinase Crystal Structure in complex with RO3201195 | | Descriptor: | Mitogen-Activated Protein Kinase 14, [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL](3-{[(2R)-2,3-DIHYDROXYPROPYL]OXY}PHENYL)METHANONE | | Authors: | Harris, S.F, Bertrand, J, Villasenor, A. | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Discovery of S-[5-Amino-1-(4-fluorophenyl)-1H-pyrazol-4-yl]-[3-(2,3-dihydroxypropoxy)phenyl]-methanone (RO3201195), and Orally Bioavailable and Highly Selective Inhibitor of p38 Map Kinase
J.Med.Chem., 49, 2006
|
|
1BT6
 
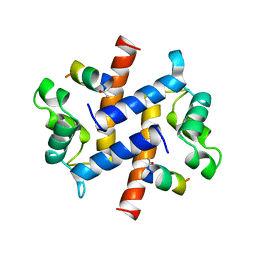 | | P11 (S100A10), LIGAND OF ANNEXIN II IN COMPLEX WITH ANNEXIN II N-TERMINUS | | Descriptor: | ANNEXIN II, S100A10 | | Authors: | Rety, S, Sopkova, J, Renouard, M, Osterloh, D, Gerke, V, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 1998-09-02 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of a complex of p11 with the annexin II N-terminal peptide.
Nat.Struct.Biol., 6, 1999
|
|
3U8U
 
 | |
6ZDJ
 
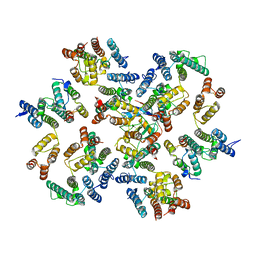 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,10) | | Descriptor: | Gag protein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-06-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WOT
 
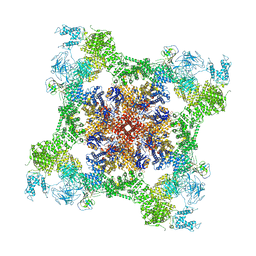 | | Cryo-EM structure of recombinant rabbit Ryanodine Receptor type 1 mutant R164C in complex with FKBP12.6 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Iyer, K.A, Hu, Y, Kurebayashi, N, Murayama, T, Samso, M. | | Deposit date: | 2020-04-25 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural mechanism of two gain-of-function cardiac and skeletal RyR mutations at an equivalent site by cryo-EM.
Sci Adv, 6, 2020
|
|
6NBS
 
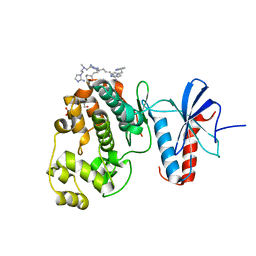 | | WT ERK2 with compound 2507-8 | | Descriptor: | (5S)-5-benzyl-4,5-dihydro-1H-imidazol-2-amine, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | Sammons, R.M, Perry, N.A, Cho, E.J, Kaoud, T.S, Zamora-Olivares, D.P, Piserchio, A, Houghten, R.A, Giulianotti, M, Li, Y, Debevec, G, Gurevich, V.V, Ghose, R, Iverson, T.M, Dalby, K.N. | | Deposit date: | 2018-12-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Class of Common Docking Domain Inhibitors That Prevent ERK2 Activation and Substrate Phosphorylation.
Acs Chem.Biol., 14, 2019
|
|
4CR4
 
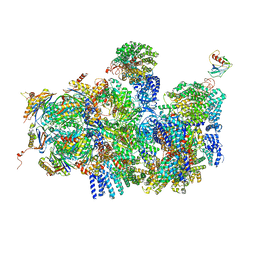 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CR2
 
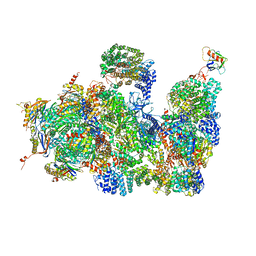 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6L2B
 
 | |
1F40
 
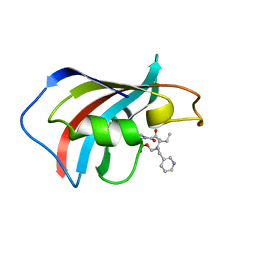 | | SOLUTION STRUCTURE OF FKBP12 COMPLEXED WITH GPI-1046, A NEUROTROPHIC LIGAND | | Descriptor: | (2S)-[3-PYRIDYL-1-PROPYL]-1-[3,3-DIMETHYL-1,2-DIOXOPENTYL]-2-PYRROLIDINECARBOXYLATE, FK506 BINDING PROTEIN (FKBP12) | | Authors: | Sich, C, Improta, S, Cowley, D.J, Guenet, C, Merly, J.P, Teufel, M, Saudek, V. | | Deposit date: | 2000-06-07 | | Release date: | 2000-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a neurotrophic ligand bound to FKBP12 and its effects on protein dynamics.
Eur.J.Biochem., 267, 2000
|
|
3ROC
 
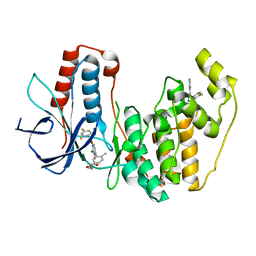 | | Crystal structure of human p38 alpha complexed with a pyrimidinone compound | | Descriptor: | 3-{5-chloro-4-[(2,4-difluorobenzyl)oxy]-6-oxopyrimidin-1(6H)-yl}-N-(2-hydroxyethyl)-4-methylbenzamide, 4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Shieh, H.-S, Xing, L. | | Deposit date: | 2011-04-25 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substituted N-aryl-6-pyrimidinones: A new class of potent, selective, and orally active p38 MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7QPD
 
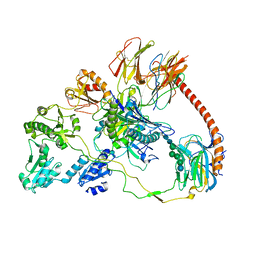 | | Structure of the human MHC I peptide-loading complex editing module | | Descriptor: | Beta-2-microglobulin, Calreticulin, HLA class I histocompatibility antigen, ... | | Authors: | Domnick, A, Susac, L, Trowitzsch, S, Thomas, C, Tampe, R. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Molecular basis of MHC I quality control in the peptide loading complex.
Nat Commun, 13, 2022
|
|
7SUV
 
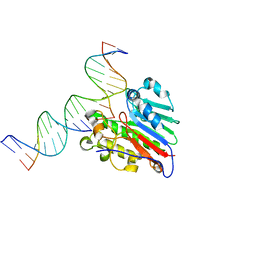 | | APE1 exonuclease substrate complex with 8oxoG opposite A | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Whitaker, A.W, Freudenthal, B.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Processing oxidatively damaged bases at DNA strand breaks by APE1.
Nucleic Acids Res., 50, 2022
|
|
4CR3
 
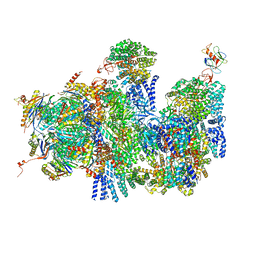 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7SVB
 
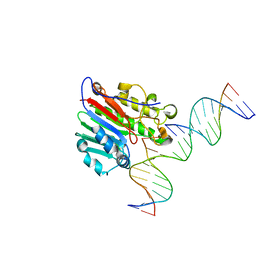 | | APE1 exonuclease substrate complex with 8oxoG opposite C | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Whitaker, A.W, Freudenthal, B.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Processing oxidatively damaged bases at DNA strand breaks by APE1.
Nucleic Acids Res., 50, 2022
|
|
3RIN
 
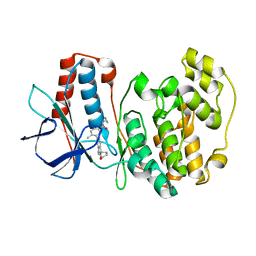 | | p38 kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-4-methyl-3-(2'-oxo-1',2'-dihydrospiro[cyclopentane-1,3'-indol]-6'-yl)benzamide | | Authors: | Segarra, V, Eastwood, P, Roca, R, Fisher, M, Lamers, M. | | Deposit date: | 2011-04-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Indolin-2-one p38(alpha) inhibitors I: design, profiling and crystallographic binding mode.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6OPI
 
 | | phosphorylated ERK2 with SCH-CPD336 | | Descriptor: | (3R)-N-[3-(2-cyclopropylpyridin-4-yl)-1H-indazol-5-yl]-3-(methoxymethyl)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}ethyl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1 | | Authors: | Vigers, G.P, Smith, D. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Activation loop dynamics are controlled by conformation-selective inhibitors of ERK2.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7ABT
 
 | |
1FKH
 
 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | Descriptor: | 1-CYCLOHEXYL-3-PHENYL-1-PROPYL-1-(3,3-DIMETHYL-1,2-DIOXYPENTYL)-2-PIPERIDINE CARBOXYLATE, FK506 BINDING PROTEIN | | Authors: | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | Deposit date: | 1993-08-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
