7ORR
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00022 | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7OPL
 
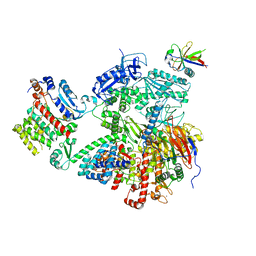 | |
7P8J
 
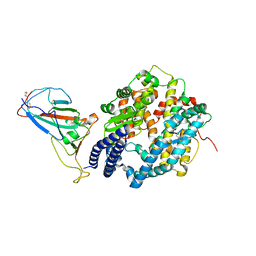 | | Receptor-binding domain (RBD) of the spike protein of the bat coronavirus RaTG13 virus in complex with the extracellular domain of human angiotensin-converting enzyme 2 (ACE2) - Crystal form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Scietti, L, Castelli, M, Faravelli, S, Clementi, N, Mancini, N, Forneris, F. | | Deposit date: | 2021-07-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (6.585 Å) | | Cite: | Constrained Evolution of SARS-CoV-2 Spike in Rhinolophus affinis Bats
To Be Published
|
|
7P8I
 
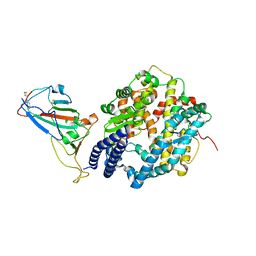 | | Receptor-binding domain (RBD) of the spike protein of the bat coronavirus RaTG13 virus in complex with the extracellular domain of human angiotensin-converting enzyme 2 (ACE2) - Crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Scietti, L, Castelli, M, Faravelli, S, Clementi, N, Mancini, N, Forneris, F. | | Deposit date: | 2021-07-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Evidence of SARS-CoV-2 Direct Evolution in R. affinis Bats Driven by Affinity and Dynamics Optimization of the Spike Protein
To Be Published
|
|
7E3O
 
 | |
7QG7
 
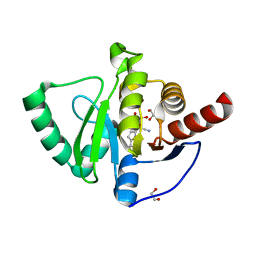 | | SARS-CoV-2 macrodomain Nsp3b bound to the remdesivir nucleoside GS-441524 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile, 1,2-ETHANEDIOL, Papain-like protease nsp3 | | Authors: | Wollenhaupt, J, Linhard, V, Sreeramulu, S, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2021-12-07 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Binding Adaptation of GS-441524 Diversifies Macro Domains and Downregulates SARS-CoV-2 de-MARylation Capacity.
J.Mol.Biol., 434, 2022
|
|
7QGI
 
 | | Crystal structure of SARS-CoV-2 NSP14 in the absence of NSP10 | | Descriptor: | PHOSPHATE ION, Proofreading exoribonuclease nsp14, ZINC ION | | Authors: | Newman, J.A, Imprachim, N, Yosaatmadja, Y, Gileadi, O. | | Deposit date: | 2021-12-08 | | Release date: | 2022-01-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.
Nucleic Acids Res., 51, 2023
|
|
7QIF
 
 | | Crystal structure of SARS-CoV-2 NSP14 in complex with 7MeGpppG. | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Newman, J.A, Imprachim, N, Yosaatmadja, Y, Gileadi, O. | | Deposit date: | 2021-12-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.
Nucleic Acids Res., 51, 2023
|
|
7QBB
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 18 | | Descriptor: | 3C-like proteinase nsp5, 7-isoquinolin-4-yl-2-phenyl-5,7-diazaspiro[3.4]octane-6,8-dione, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-11-18 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7QCT
 
 | | PDZ2 of LNX2 with SARS-CoV-2_E PBM complex | | Descriptor: | Envelope small membrane protein, Ligand of Numb protein X 2 | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
7QCR
 
 | | MLLT4/Afadin PDZ domain in complex with the C-terminal peptide from protein E of SARS-CoV-2 | | Descriptor: | Afadin, Envelope small membrane protein, SULFATE ION | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
7QCS
 
 | | PALS1/MPP5 PDZ domain in complex with SARS-CoV-2_E PBM peptide | | Descriptor: | Envelope small membrane protein, Protein PALS1 | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
7QLF
 
 | |
7PHG
 
 | | MaP OF P5C3RBD Interface | | Descriptor: | Heavy ChaIn variable, Light ChaIn, Surface glycoprotein | | Authors: | Perez, L. | | Deposit date: | 2021-08-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A highly potent antibody effective against SARS-CoV-2 variants of concern.
Cell Rep, 37, 2021
|
|
7X7T
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zheng, Q, Li, S, Zhang, T, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7V
 
 | | Cryo-EM structure of SARS-CoV spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7U
 
 | | Cryo-EM structure of SARS-CoV-2 Delta variant spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XA7
 
 | | Crystal structure of SARS-CoV-2 receptor-binding domain in complex with intermediate horseshoe bat ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Tang, L.F, Zhang, D, Han, P, Qi, J.X. | | Deposit date: | 2022-03-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural basis of SARS-CoV-2 and its variants binding to intermediate horseshoe bat ACE2.
Int J Biol Sci, 18, 2022
|
|
7X2H
 
 | |
7X63
 
 | | SARS-CoV-2-Beta-RBD and BD-236-GWP/P-VK antibody complex | | Descriptor: | BD-236 Fab heavy chain, BD-236 Fab light chain, Spike protein S1 | | Authors: | Shi, R, Wang, Y, Wu, Z, Han, X, Yan, J. | | Deposit date: | 2022-03-06 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | SARS-CoV-2-Beta-RBD and BD-236-GWP/P-VK antibody complex
To Be Published
|
|
7X66
 
 | | SARS-CoV-2-Omicron-RBD and BD-236-GWP/P-VK antibody complex | | Descriptor: | BD-236 Fab heavy chain, BD-236 Fab light chain, Spike protein S1 | | Authors: | Shi, R, Wang, Y, Wu, Z, Han, X, Yan, J. | | Deposit date: | 2022-03-06 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2-Omicron-RBD and BD-236-GWP/P-VK antibody complex
To Be Published
|
|
7X93
 
 | |
7X95
 
 | |
7X94
 
 | |
7X96
 
 | |
