9E55
 
 | |
9ERH
 
 | |
1K9M
 
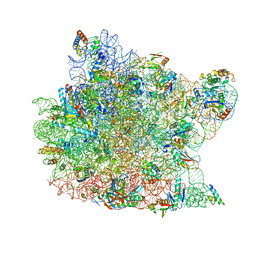 | | Co-crystal structure of tylosin bound to the 50S ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-10-29 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
1KE5
 
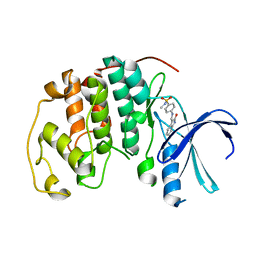 | | CDK2 complexed with N-methyl-4-{[(2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]amino}benzenesulfonamide | | Descriptor: | Cell division protein kinase 2, N-METHYL-4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KEY
 
 | | Crystal Structure of Mouse Testis/Brain RNA-binding Protein (TB-RBP) | | Descriptor: | translin | | Authors: | Pascal, J.M, Hart, P.J, Hecht, N.B, Robertus, J.D. | | Deposit date: | 2001-11-19 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of TB-RBP, a Novel RNA-binding and Regulating Protein
J.Mol.Biol., 319, 2002
|
|
1KF8
 
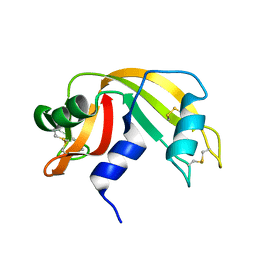 | | Atomic resolution structure of RNase A at pH 8.8 | | Descriptor: | pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8EOW
 
 | |
1KEA
 
 | | STRUCTURE OF A THERMOSTABLE THYMINE-DNA GLYCOSYLASE | | Descriptor: | ACETATE ION, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Mol, C.D, Arvai, A.S, Begley, T.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 2001-11-14 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of a thermostable thymine-DNA glycosylase: evidence for base twisting to remove mismatched normal DNA bases.
J.Mol.Biol., 315, 2002
|
|
8EFO
 
 | | PZM21-bound mu-opioid receptor-Gi complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
1KEH
 
 | | Precursor structure of cephalosporin acylase | | Descriptor: | precursor of cephalosporin acylase | | Authors: | Kim, Y, Kim, S. | | Deposit date: | 2001-11-16 | | Release date: | 2002-05-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Precursor structure of cephalosporin acylase. Insights into autoproteolytic activation in a new N-terminal hydrolase family
J.Biol.Chem., 277, 2002
|
|
1KFI
 
 | | Crystal Structure of the Exocytosis-Sensitive Phosphoprotein, pp63/Parafusin (phosphoglucomutase) from Paramecium | | Descriptor: | SULFATE ION, ZINC ION, phosphoglucomutase 1 | | Authors: | Mueller, S, Diederichs, K, Breed, J, Kissmehl, R, Hauser, K, Plattner, H, Welte, W. | | Deposit date: | 2001-11-21 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure analysis of the exocytosis-sensitive phosphoprotein, pp63/parafusin (phosphoglucomutase), from Paramecium reveals significant conformational variability.
J.Mol.Biol., 315, 2002
|
|
1KFZ
 
 | | Solution Structure of C-terminal Sem-5 SH3 Domain (Ensemble of 16 Structures) | | Descriptor: | SEX MUSCLE ABNORMAL PROTEIN 5 | | Authors: | Ferreon, J.C, Volk, D.E, Luxon, B.A, Gorenstein, D, Hilser, V.J. | | Deposit date: | 2001-11-24 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics and Thermodynamics of the Native State Ensemble
of Sem-5 C-Terminal SH3 Domain
Biochemistry, 42, 2003
|
|
1KGO
 
 | | R2F from Corynebacterium Ammoniagenes in its reduced, Fe containing, form | | Descriptor: | FE (II) ION, Ribonucleotide reductase protein R2F | | Authors: | Hogbom, M, Huque, Y, Sjoberg, B.M, Nordlund, P. | | Deposit date: | 2001-11-28 | | Release date: | 2001-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the di-iron/radical protein of ribonucleotide reductase from Corynebacterium ammoniagenes.
Biochemistry, 41, 2002
|
|
4X3G
 
 | | Crystal structure of SIAH1 SINA domain in complex with a USP19 peptide | | Descriptor: | E3 ubiquitin-protein ligase SIAH1, Ubiquitin carboxyl-terminal hydrolase 19, ZINC ION | | Authors: | Walker, J.R, Dong, A, Zhang, Q, Huang, X, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-11-28 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of SIAH1 SINA domain in complex with a USP19 peptide
To be published
|
|
4X2T
 
 | | X-ray crystal structure of the orally available aminopeptidase inhibitor, Tosedostat, bound to the M17 Leucyl Aminopeptidase from P. falciparum | | Descriptor: | (2S)-({(2R)-2-[(1S)-1-hydroxy-2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}amino)(phenyl)ethanoic acid, CARBONATE ION, M17 leucyl aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | X-ray crystal structures of an orally available aminopeptidase inhibitor, Tosedostat, bound to anti-malarial drug targets PfA-M1 and PfA-M17.
Proteins, 83, 2015
|
|
1KHY
 
 | |
6UIG
 
 | |
1KI9
 
 | |
4X4O
 
 | |
1KJ5
 
 | | Solution Structure of Human beta-defensin 1 | | Descriptor: | BETA-DEFENSIN 1 | | Authors: | Schibli, D.J, Hunter, H.N, Aseyev, V, Starner, T.D, Wiencek, J.M, McCray Jr, P.B, Tack, B.F, Vogel, H.J. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-20 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus.
J.Biol.Chem., 277, 2002
|
|
1KFH
 
 | |
8EFL
 
 | | SR17018-bound mu-opioid receptor-Gi complex | | Descriptor: | 5,6-dichloro-1-{1-[(4-chlorophenyl)methyl]piperidin-4-yl}-1,3-dihydro-2H-benzimidazol-2-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
4X6A
 
 | | Crystal structure of yeast RNA polymerase II encountering oxidative Cyclopurine DNA lesions | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2014-12-07 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1KG2
 
 | | Crystal structure of the core fragment of MutY from E.coli at 1.2A resolution | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KJO
 
 | |
