7COQ
 
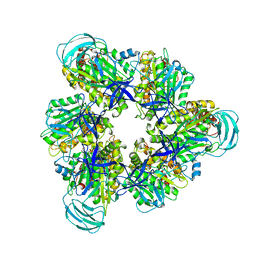 | |
7CP3
 
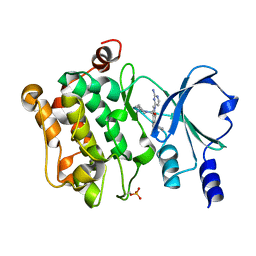 | | Crystal Structure of PAK4 in complex with inhibitor 47 | | Descriptor: | Serine/threonine-protein kinase PAK 4, [(3R)-3-azanylpiperidin-1-yl]-[1-(2-azanylpyrimidin-4-yl)-6-[2-(1-oxidanylcyclohexyl)ethynyl]indol-3-yl]methanone | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2020-08-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of 6-ethynyl-1H-indole-3-carboxamide Derivatives as Highly Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors
to be published
|
|
7JNT
 
 | |
7JNY
 
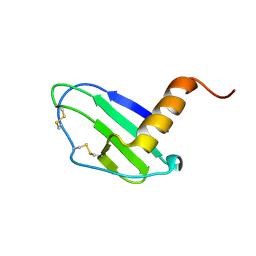 | | Crystal structure of CXCL13 | | Descriptor: | C-X-C motif chemokine 13 | | Authors: | Rosenberg Jr, E.M, Rajasekaran, D, Murphy, J.W, Pantouris, G, Lolis, E.J. | | Deposit date: | 2020-08-05 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The N-terminal length and side-chain composition of CXCL13 affect crystallization, structure and functional activity.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZZ8
 
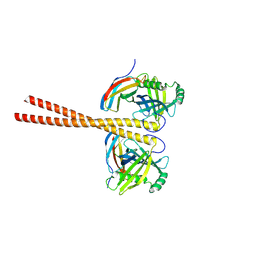 | | MB_CRS6-15 bound to CrSAS-6_6HR | | Descriptor: | Centriole protein, Protein B | | Authors: | Hatzopoulos, G.N, Kukenshoner, T, Banterle, N, Favez, T, Fluckiger, I, Hantschel, O, Gonczy, P. | | Deposit date: | 2020-08-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | Tuning SAS-6 architecture with monobodies impairs distinct steps of centriole assembly.
Nat Commun, 12, 2021
|
|
7JNE
 
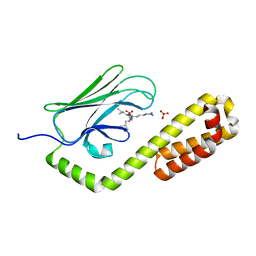 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RGSQLRIASR | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZZE
 
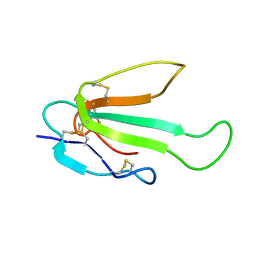 | |
7JNI
 
 | | Crystal structure of the angiotensin II type 2 receptoror (AT2R) in complex with EMA401 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FORMIC ACID, HEXANE-1,6-DIOL, ... | | Authors: | Cherezov, V, Shaye, H, Han, G.W. | | Deposit date: | 2020-08-04 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of the angiotensin II type 2 receptor AT 2 R is a novel therapeutic strategy for glioblastoma.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7JN8
 
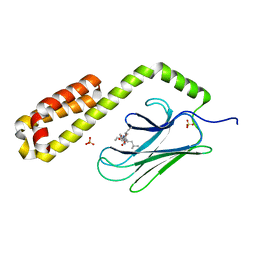 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RGNTLVIVSR | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7COO
 
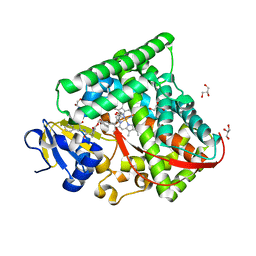 | |
7CON
 
 | |
7JN9
 
 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide QEHTGSQLRIAAYGP | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZZF
 
 | |
6ZZ2
 
 | | Cocktail experiment E: fragments 52, 58, and 63 at 90 mM concentration in complex with Endothiapepsin | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | Authors: | Hassaan, E, Klebe, G, Heine, A, Schiebel, J, Koester, H. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14885437 Å) | | Cite: | Cocktail experiment E: fragments 52, 58, and 63 at 90 mM concentration in complex with Endothiapepsin
To Be Published
|
|
6ZYU
 
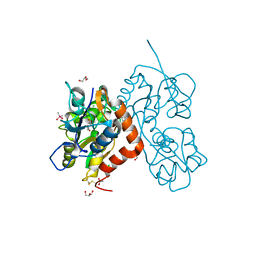 | | Structure of the GluA2 ligand-binding domain (L483Y-N754S) in complex with glutamate and BPAM549 | | Descriptor: | (4~{R})-4-cyclopropyl-7-fluoranyl-3,4-dihydro-2~{H}-thiochromene 1,1-dioxide, (4~{S})-4-cyclopropyl-7-fluoranyl-3,4-dihydro-2~{H}-thiochromene 1,1-dioxide, ACETATE ION, ... | | Authors: | Dorosz, J, Christensen, K.M, Kastrup, J.S. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Thiochroman Dioxide Analogues of Benzothiadiazine Dioxides as New Positive Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors.
Acs Chem Neurosci, 12, 2021
|
|
7JN0
 
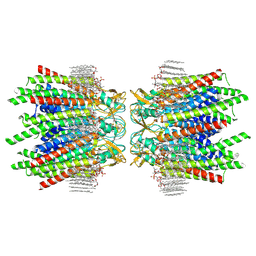 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 2 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-08-03 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JN4
 
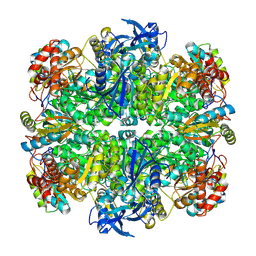 | | Rubisco in the apo state | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain 2, chloroplastic | | Authors: | Matthies, D, Jonikas, M.C, He, S. | | Deposit date: | 2020-08-03 | | Release date: | 2020-11-18 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | The structural basis of Rubisco phase separation in the pyrenoid.
Nat.Plants, 6, 2020
|
|
7JN1
 
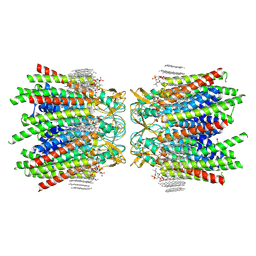 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-08-03 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
6ZYW
 
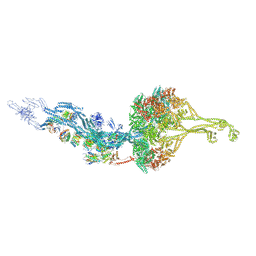 | | Outer Dynein Arm-Shulin complex - overall structure (Tetrahymena thermophila) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Dynein heavy chain, ... | | Authors: | Mali, G.R, Abid Ali, F, Lau, C.K, Carter, A.P. | | Deposit date: | 2020-08-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (8.78 Å) | | Cite: | Shulin packages axonemal outer dynein arms for ciliary targeting.
Science, 371, 2021
|
|
7CNN
 
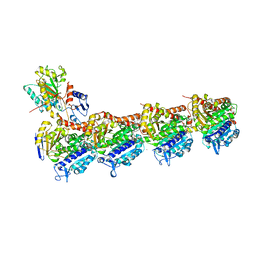 | | vinorelbine in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, Y.X, Wu, C.Y. | | Deposit date: | 2020-08-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The high-resolution X-ray structure of vinca-domain inhibitors of microtubules provides a rational approach for drug design.
Febs Lett., 595, 2021
|
|
7CNO
 
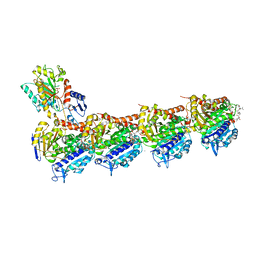 | | Phomopsin A in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, C.Y, Wang, Y.X. | | Deposit date: | 2020-08-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The high-resolution X-ray structure of vinca-domain inhibitors of microtubules provides a rational approach for drug design.
Febs Lett., 595, 2021
|
|
7CNM
 
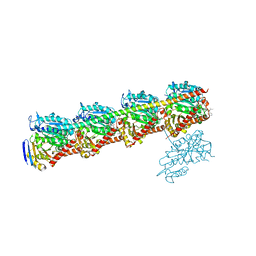 | | YDX in complex with tubulin | | Descriptor: | (2~{S},4~{S})-4-[[2-[(1~{R},3~{R})-1-acetyloxy-4-methyl-3-[[(2~{S},3~{S})-3-methyl-2-[[(2~{R})-1-methylpiperidin-2-yl]carbonylamino]pentanoyl]amino]pentyl]-1,3-thiazol-4-yl]carbonylamino]-5-cyclohexyl-2-methyl-pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y.X, Wu, C.Y. | | Deposit date: | 2020-08-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The high-resolution X-ray structure of vinca-domain inhibitors of microtubules provides a rational approach for drug design.
Febs Lett., 595, 2021
|
|
7JMM
 
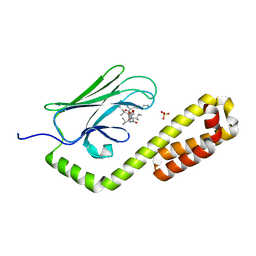 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RAKNIILLSR | | Descriptor: | Alkaline phosphatase, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2020-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JMU
 
 | | Hen egg-white lysozyme with ionic liquid ethylammonium nitrate | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Han, Q, Darmanin, C, Smith, K, Greaves, T, Drummond, C. | | Deposit date: | 2020-08-02 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Lysozyme conformational changes with ionic liquids: Spectroscopic, small angle x-ray scattering and crystallographic study.
J Colloid Interface Sci, 585, 2021
|
|
6ZYM
 
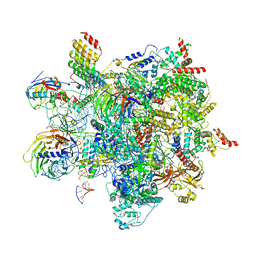 | | Human C Complex Spliceosome - High-resolution CORE | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Corepressor interacting with RBPJ 1, ... | | Authors: | Bertram, K, Kastner, B. | | Deposit date: | 2020-08-02 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insights into the Roles of Metazoan-Specific Splicing Factors in the Human Step 1 Spliceosome.
Mol.Cell, 80, 2020
|
|
