4LHL
 
 | |
2EIN
 
 | | Zinc ion binding structure of bovine heart cytochrome C oxidase in the fully oxidized state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Hirata, K, Shinzawa-Itoh, K, Yoko-o, S, Yamashita, E, Aoyama, H, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2007-03-13 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A histidine residue acting as a controlling site for dioxygen reduction and proton pumping by cytochrome c oxidase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4C0J
 
 | | Crystal structure of Drosophila Miro EF hand and cGTPase domains in the apo state (Apo-MiroS) | | Descriptor: | L-HOMOSERINE, MITOCHONDRIAL RHO GTPASE, SODIUM ION, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Wawrzak, Z, Chakravarthy, S, Landahl, E.C, Freymann, D.M, Rice, S.E. | | Deposit date: | 2013-08-05 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Coupling of the EF Hand and C-Terminal Gtpase Domains in the Mitochondrial Protein Miro.
Embo Rep., 14, 2013
|
|
2VWN
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-Chloro-thiophene-2-carboxylic acid ((3S,4S)-1-{[2-fluoro-4-(2-oxo-2H-pyridin-1-yl)-phenylcarbamoyl]-methyl}-4-hydroxy-pyrrolidin-3-yl)-amide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
4C0K
 
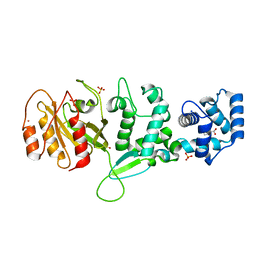 | | Crystal structure of Drosophila Miro EF hand and cGTPase domains bound to one calcium ion (Ca-MiroS) | | Descriptor: | CALCIUM ION, L-HOMOSERINE, MITOCHONDRIAL RHO GTPASE, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Wawrzak, Z, Chakravarthy, S, Landahl, E.C, Freymann, D.M, Rice, S.E. | | Deposit date: | 2013-08-05 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural Coupling of the EF Hand and C-Terminal Gtpase Domains in the Mitochondrial Protein Miro.
Embo Rep., 14, 2013
|
|
4C0L
 
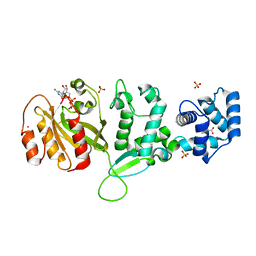 | | Crystal structure of Drosophila Miro EF hand and cGTPase domains bound to one magnesium ion and Mg:GDP (MgGDP-MiroS) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, L-HOMOSERINE, MAGNESIUM ION, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Wawrzak, Z, Chakravarthy, S, Landahl, E.C, Freymann, D.M, Rice, S.E. | | Deposit date: | 2013-08-05 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Coupling of the EF Hand and C-Terminal Gtpase Domains in the Mitochondrial Protein Miro.
Embo Rep., 14, 2013
|
|
3G1N
 
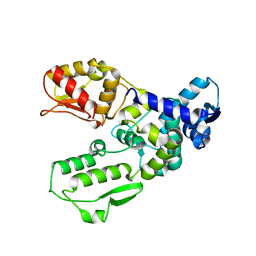 | | Catalytic domain of the human E3 ubiquitin-protein ligase HUWE1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1, SODIUM ION | | Authors: | Walker, J.R, Qiu, L, Li, Y, Davis, T, Tempel, W, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hect Domain of Human HUWE1/MULE
To be Published
|
|
3P2Y
 
 | |
3FZQ
 
 | |
3GED
 
 | | Fingerprint and Structural Analysis of a Apo SCOR enzyme from Clostridium thermocellum | | Descriptor: | GLYCEROL, SODIUM ION, Short-chain dehydrogenase/reductase SDR, ... | | Authors: | Huether, R, Liu, Z.J, Xu, H, Wang, B.C, Pletnev, V, Mao, Q, Umland, T, Duax, W. | | Deposit date: | 2009-02-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Sequence fingerprint and structural analysis of the SCOR enzyme A3DFK9 from Clostridium thermocellum.
Proteins, 78, 2010
|
|
4BY5
 
 | |
3P9F
 
 | | Urate oxidase-azaxanthine-azide ternary complex | | Descriptor: | 8-AZAXANTHINE, AZIDE ION, SODIUM ION, ... | | Authors: | Gabison, L, Colloc'H, N, El Hajji, M, Castro, B, Chiadmi, M, Prange, T. | | Deposit date: | 2010-10-17 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Azide and cyanide have different inhibition modes of urate oxidase
To be Published
|
|
4C3W
 
 | | Vanadium(IV)-Picolinate Complexed with Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, LYSOZYME C, ... | | Authors: | Santos, M.F.A, Correia, I, Oliveira, A.R, Garribba, E, Pessoa, J.C, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2013-08-28 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Vanadium Complexes as Prospective Therapeutics: Structural Characterization of a V(Iv) Lysozyme Adduct
Eur.J.Inorg.Chem., 21, 2014
|
|
4LDE
 
 | | Structure of beta2 adrenoceptor bound to BI167107 and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid Antibody Fragment, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
4N49
 
 | | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 Protein in complex with m7GpppG and SAM | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Smietanski, M, Werener, M, Purta, E, Kaminska, K.H, Stepinski, J, Darzynkiewicz, E, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation.
Nat Commun, 5, 2014
|
|
2V9V
 
 | | Crystal Structure of Moorella thermoacetica SelB(377-511) | | Descriptor: | CHLORIDE ION, SELENOCYSTEINE-SPECIFIC ELONGATION FACTOR, SODIUM ION | | Authors: | Ganichkin, O, Wahl, M.C. | | Deposit date: | 2007-08-27 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Conformational Switches in Winged-Helix Domains 1 and 2 of Bacterial Translation Elongation Factor Selb.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4ES8
 
 | | Crystal Structure of the adhesin domain of Epf from Streptococcus pyogenes in P212121 | | Descriptor: | ACETATE ION, Epf, GLYCEROL, ... | | Authors: | Linke, C, Siemens, N, Kreikemeyer, B, Baker, E.N. | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The Extracellular Protein Factor Epf from Streptococcus pyogenes Is a Cell Surface Adhesin That Binds to Cells through an N-terminal Domain Containing a Carbohydrate-binding Module.
J.Biol.Chem., 287, 2012
|
|
4EUF
 
 | | Crystal structure of Clostridium acetobutulicum trans-2-enoyl-CoA reductase in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative reductase CA_C0462, SODIUM ION | | Authors: | Hu, K, Zhao, M, Zhang, T, Yang, S, Ding, J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of trans-2-enoyl-CoA reductases from Clostridium acetobutylicum and Treponema denticola: insights into the substrate specificity and the catalytic mechanism
Biochem.J., 449, 2013
|
|
1S6Q
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R147681 | | Descriptor: | 4-[4-(2,4,6-TRIMETHYL-PHENYLAMINO)-PYRIMIDIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
4N9V
 
 | | High resolution x-ray structure of urate oxidase in complex with 8-azaxanthine | | Descriptor: | 8-AZAXANTHINE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Oksanen, E, Blakeley, M.P, Budayova-Spano, M. | | Deposit date: | 2013-10-21 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The neutron structure of urate oxidase resolves a long-standing mechanistic conundrum and reveals unexpected changes in protonation.
Plos One, 9, 2014
|
|
1S3P
 
 | |
4EEK
 
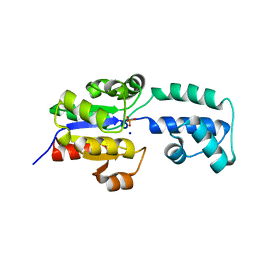 | | Crystal structure of HAD FAMILY HYDROLASE DR_1622 from Deinococcus radiodurans R1 (TARGET EFI-501256) With bound phosphate and sodium | | Descriptor: | Beta-phosphoglucomutase-related protein, PHOSPHATE ION, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of HAD HYDROLASE DR_1622 Deinococcus radiodurans R1 (TARGET EFI-501256)
To be Published
|
|
4MV2
 
 | | Crystal structure of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 | | Authors: | Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Thomas, M.G, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-02 | | Last modified: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
4EEB
 
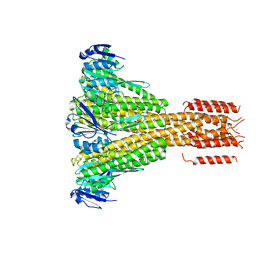 | | CorA coiled-coil mutant under Mg2+ absence | | Descriptor: | CESIUM ION, Magnesium transport protein CorA, SODIUM ION | | Authors: | Pfoh, R, Pai, E.F. | | Deposit date: | 2012-03-28 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural asymmetry in the magnesium channel CorA points to sequential allosteric regulation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3FYH
 
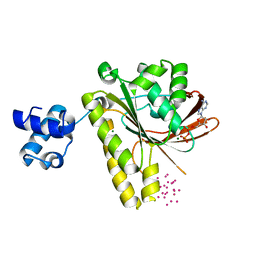 | | Recombinase in complex with ADP and metatungstate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair and recombination protein radA, MAGNESIUM ION, ... | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2009-01-22 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an archaeal Rad51 homologue in complex with a metatungstate inhibitor.
Biochemistry, 48, 2009
|
|
