6GUE
 
 | | CDK2/CyclinA in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
5UVK
 
 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
Iucrj, 4, 2017
|
|
1K06
 
 | | Crystallographic Binding Study of 100 mM N-benzoyl-N'-beta-D-glucopyranosyl urea to glycogen phosphorylase b | | Descriptor: | Glycogen Phosphorylase, N-[(phenylcarbonyl)carbamoyl]-beta-D-glucopyranosylamine, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Kosmopoulou, M, Zographos, S.E, Leonidas, D.D, Chrysina, E.D, Somsak, L, Nagy, V, Praly, J.P, Docsa, T, Toth, B, Gergely, P. | | Deposit date: | 2001-09-18 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of N-acetyl-N '-beta-D-glucopyranosyl urea and N-benzoyl-N '-beta-D-glucopyranosyl urea to glycogen phosphorylase b: kinetic and crystallographic studies.
Eur.J.Biochem., 269, 2002
|
|
2HRL
 
 | | Siglec-7 in complex with GT1b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ETHYL-TRIMETHYL-SILANE, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-[2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Attrill, H, Imamura, A, Sharma, R.S, Kiso, M, Crocker, P.R, van Aalten, D.M.F. | | Deposit date: | 2006-07-20 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Siglec-7 Undergoes a Major Conformational Change When Complexed with the {alpha}(2,8)-Disialylganglioside GT1b
J.Biol.Chem., 281, 2006
|
|
1D7N
 
 | | SOLUTION STRUCTURE ANALYSIS OF THE MASTOPARAN WITH DETERGENTS | | Descriptor: | PROTEIN (WASP VENOM PEPTIDE (MASTOPARAN)) | | Authors: | Hori, Y, Demura, M, Iwadate, M, Niidome, T, Aoyagi, H, Asakura, T. | | Deposit date: | 1999-10-19 | | Release date: | 2001-06-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Interaction of mastoparan with membranes studied by 1H-NMR spectroscopy in detergent micelles and by solid-state 2H-NMR and 15N-NMR spectroscopy in oriented lipid bilayers.
Eur.J.Biochem., 268, 2001
|
|
1LO5
 
 | | Crystal structure of the D227A variant of Staphylococcal enterotoxin A in complex with human MHC class II | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DR-1 beta chain, ... | | Authors: | Petersson, K, Thunnissen, M, Forsberg, G, Walse, B. | | Deposit date: | 2002-05-06 | | Release date: | 2002-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of a SEA Variant in Complex with MHC Class II Reveals the Ability of SEA to Crosslink MHC Molecules
Structure, 10, 2002
|
|
1KZA
 
 | | Complex of MBP-C and Man-a13-Man | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN C, ... | | Authors: | Ng, K.K, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
1L9Y
 
 | | FEZ-1-Y228A, A Mutant of the Metallo-beta-lactamase from Legionella gormanii | | Descriptor: | CHLORIDE ION, FEZ-1 b-lactamase, GLYCEROL, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Galleni, M, Dideberg, O. | | Deposit date: | 2002-03-27 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Three-dimensional Structure of FEZ-1, a Monomeric Subclass B3 Metallo-beta-lactamase
from Fluoribacter gormanii, in Native Form and in Complex with -Captopril
J.Mol.Biol., 325, 2003
|
|
5UVL
 
 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
IUCrJ, 4, 2017
|
|
1K7A
 
 | | Ets-1(331-440)+GGAG duplex | | Descriptor: | C-ets-1 Protein, DNA (5'-D(*CP*AP*CP*AP*TP*CP*TP*CP*CP*GP*GP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*CP*CP*GP*GP*AP*GP*AP*TP*GP*T)-3') | | Authors: | Garvie, C.W, Hagman, J, Wolberger, C. | | Deposit date: | 2001-10-18 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of Ets-1/Pax5 complex formation on DNA.
Mol.Cell, 8, 2001
|
|
2H4F
 
 | | Sir2-p53 peptide-NAD+ | | Descriptor: | Cellular tumor antigen p53, NAD-dependent deacetylase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hoff, K.G, Avalos, J.L, Sens, K, Wolberger, C. | | Deposit date: | 2006-05-24 | | Release date: | 2006-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the Sirtuin Mechanism from Ternary Complexes Containing NAD(+) and Acetylated Peptide.
Structure, 14, 2006
|
|
6GUC
 
 | | CDK2/CyclinA in complex with SU9516 | | Descriptor: | (3Z)-3-(1H-IMIDAZOL-5-YLMETHYLENE)-5-METHOXY-1H-INDOL-2(3H)-ONE, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
2GPR
 
 | | GLUCOSE PERMEASE IIA FROM MYCOPLASMA CAPRICOLUM | | Descriptor: | GLUCOSE-PERMEASE IIA COMPONENT | | Authors: | Huang, K, Herzberg, O. | | Deposit date: | 1998-05-19 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A promiscuous binding surface: crystal structure of the IIA domain of the glucose-specific permease from Mycoplasma capricolum.
Structure, 6, 1998
|
|
2GN8
 
 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADP and UDP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-GlcNAc C6 dehydratase, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
1K61
 
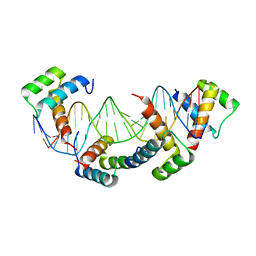 | | MATALPHA2 HOMEODOMAIN BOUND TO DNA | | Descriptor: | 5'-D(*(5IU)P*GP*CP*GP*TP*GP*TP*AP*AP*AP*TP*GP*AP*AP*TP*TP*AP*CP*AP*TP*G)-3', 5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*CP*AP*TP*TP*TP*AP*CP*AP*CP*GP*C)-3', Mating-type protein alpha-2 | | Authors: | Aishima, J, Gitti, R.K, Noah, J.E, Gan, H.H, Schlick, T, Wolberger, C. | | Deposit date: | 2001-10-14 | | Release date: | 2002-12-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Hoogsteen base pair embedded in undistorted B-DNA
NUCLEIC ACIDS RES., 30, 2002
|
|
1K78
 
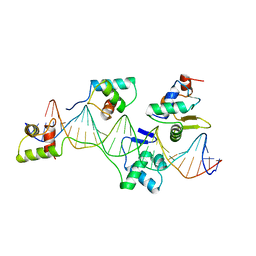 | | Pax5(1-149)+Ets-1(331-440)+DNA | | Descriptor: | C-ets-1 Protein, Paired Box Protein Pax5, Pax5/Ets Binding Site on the mb-1 promoter | | Authors: | Garvie, C.W, Hagman, J, Wolberger, C. | | Deposit date: | 2001-10-18 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural studies of Ets-1/Pax5 complex formation on DNA.
Mol.Cell, 8, 2001
|
|
2H1V
 
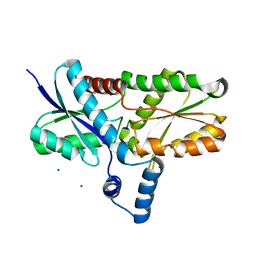 | | Crystal structure of the Lys87Ala mutant variant of Bacillus subtilis ferrochelatase | | Descriptor: | Ferrochelatase, MAGNESIUM ION | | Authors: | Hansson, M.D, Karlberg, T, Arys Rahardja, M, Al-Karadaghi, S, Hansson, M. | | Deposit date: | 2006-05-17 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Amino Acid Residues His183 and Glu264 in Bacillus subtilis Ferrochelatase Direct and Facilitate the Insertion of Metal Ion into Protoporphyrin IX
Biochemistry, 46, 2007
|
|
2H2I
 
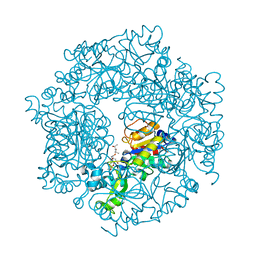 | | The Structural basis of Sirtuin Substrate Affinity | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, NAD-dependent deacetylase, ZINC ION | | Authors: | Cosgrove, M.S, Wolberger, C. | | Deposit date: | 2006-05-18 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of sirtuin substrate affinity
Biochemistry, 45, 2006
|
|
1CDZ
 
 | | BRCT DOMAIN FROM DNA-REPAIR PROTEIN XRCC1 | | Descriptor: | PROTEIN (DNA-REPAIR PROTEIN XRCC1) | | Authors: | Zhang, X, Morera, S, Bates, P, Whitehead, P, Coffer, A, Hainbucher, K, Nash, R, Sternberg, M, Lindahl, T, Freemont, P. | | Deposit date: | 1999-03-04 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an XRCC1 BRCT domain: a new protein-protein interaction module.
EMBO J., 17, 1998
|
|
6GUK
 
 | | CDK2 in complex with CGP74514A | | Descriptor: | Cyclin-dependent kinase 2, ~{N}2-[(1~{R},2~{S})-2-azanylcyclohexyl]-~{N}6-(3-chlorophenyl)-9-ethyl-purine-2,6-diamine | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
2H2G
 
 | |
1KZE
 
 | | Complex of MBP-C and bivalent Man-terminated glycopeptide | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN C, ... | | Authors: | Ng, K.K, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
1COS
 
 | | CRYSTAL STRUCTURE OF A SYNTHETIC TRIPLE-STRANDED ALPHA-HELICAL BUNDLE | | Descriptor: | COILED SERINE | | Authors: | Lovejoy, B, Choe, S, Cascio, D, Mcrorie, D.K, Degrado, W, Eisenberg, D. | | Deposit date: | 1993-01-22 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a synthetic triple-stranded alpha-helical bundle.
Science, 259, 1993
|
|
1LU4
 
 | | 1.1 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF A SECRETED MYCOBACTERIUM TUBERCULOSIS DISULFIDE OXIDOREDUCTASE HOMOLOGOUS TO E. COLI DSBE: IMPLICATIONS FOR FUNCTIONS | | Descriptor: | SOLUBLE SECRETED ANTIGEN MPT53 | | Authors: | Goulding, C.W, Apostol, M.I, Gleiter, S, Parseghian, A, Bardwell, J, Gennaro, M, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-21 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Gram-positive DsbE Proteins Function Differently from Gram-negative DsbE Homologs: A STRUCTURE TO FUNCTION ANALYSIS OF DsbE FROM MYCOBACTERIUM TUBERCULOSIS.
J.Biol.Chem., 279, 2004
|
|
1L2Q
 
 | | Crystal Structure of the Methanosarcina barkeri Monomethylamine Methyltransferase (MtmB) | | Descriptor: | AMMONIUM ION, monomethylamine methyltransferase | | Authors: | Hao, B, Gong, W, Ferguson, T.K, James, C.M, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2002-02-24 | | Release date: | 2002-06-05 | | Last modified: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A new UAG-encoded residue in the structure of a methanogen methyltransferase.
Science, 296, 2002
|
|
