6PQ3
 
 | | Crystal structure of GDP-bound KRAS with ten residues long internal tandem duplication in the switch II region | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Dharmaiah, S, Chan, A.H, Tran, T.H, Simanshu, D.K. | | Deposit date: | 2019-07-08 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | RASinternal tandem duplication disrupts GTPase-activating protein (GAP) binding to activate oncogenic signaling.
J.Biol.Chem., 295, 2020
|
|
3KQ7
 
 | |
3KSW
 
 | | Crystal structure of sterol 14alpha-demethylase (CYP51) from Trypanosoma cruzi in complex with an inhibitor VNF ((4-(4-chlorophenyl)-N-[2-(1H-imidazol-1-yl)-1-phenylethyl]benzamide) | | Descriptor: | 4'-chloro-N-[(1R)-2-(1H-imidazol-1-yl)-1-phenylethyl]biphenyl-4-carboxamide, PROTOPORPHYRIN IX CONTAINING FE, Sterol 14-alpha demethylase | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Anderson, S, Wawrzak, Z, Waterman, M.R. | | Deposit date: | 2009-11-23 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural Insights into Inhibition of Sterol 14{alpha}-Demethylase in the Human Pathogen Trypanosoma cruzi.
J.Biol.Chem., 285, 2010
|
|
4XH0
 
 | | Structure of C. glabrata Hrr25 bound to ADP (SO4 condition) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, Similar to uniprot|P29295 Saccharomyces cerevisiae YPL204w HRR25 | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-01-04 | | Release date: | 2016-01-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
4XIC
 
 | | ANTPHD WITH 15BP di-thioate modified DNA DUPLEX | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*(C2S)P*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2015-01-06 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Entropic Enhancement of Protein-DNA Affinity by Oxygen-to-Sulfur Substitution in DNA Phosphate.
Biophys.J., 109, 2015
|
|
3KUO
 
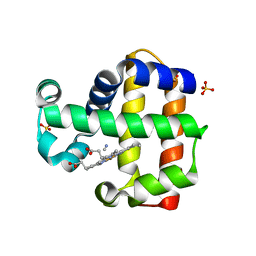 | | X-ray structure of the metcyano form of dehaloperoxidase from amphitrite ornata: evidence for photoreductive lysis of iron-cyanide bond | | Descriptor: | CYANIDE ION, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Serrano, V.S, Chen, Z, Gaff, J.F, Rose, R, Franzen, S. | | Deposit date: | 2009-11-27 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | X-ray structure of the metcyano form of dehaloperoxidase from Amphitrite ornata: evidence for photoreductive dissociation of the iron-cyanide bond.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4XOZ
 
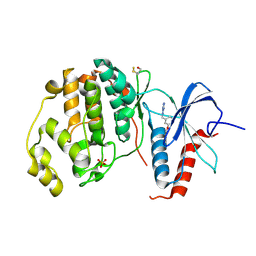 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, N~1~-[3-(benzyloxy)benzyl]-1H-tetrazole-1,5-diamine, SULFATE ION | | Authors: | Gelin, M, Allemand, F, Labesse, G, Guichou, J.F. | | Deposit date: | 2015-01-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8U1C
 
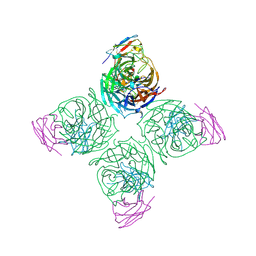 | |
4XPC
 
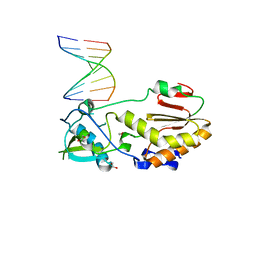 | |
6PQ0
 
 | | LCP-embedded Proteinase K treated with MPD | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Bu, G, Zhu, L, Jing, L, Shi, D, Gonen, T, Liu, W, Nannenga, B.L. | | Deposit date: | 2019-07-08 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | Structure Determination from Lipidic Cubic Phase Embedded Microcrystals by MicroED.
Structure, 28, 2020
|
|
4XJ0
 
 | | Crystal structure of ERK2 in complex with an inhibitor 14K | | Descriptor: | 1-[(1S)-1-(4-chloro-3-fluorophenyl)-2-hydroxyethyl]-4-[2-(tetrahydro-2H-pyran-4-ylamino)pyrimidin-4-yl]pyridin-2(1H)-one, Mitogen-activated protein kinase 1 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2015-01-08 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of highly potent, selective, and efficacious small molecule inhibitors of ERK1/2.
J.Med.Chem., 58, 2015
|
|
8TRN
 
 | | Actin 1 from T. gondii in filaments bound to MgADP and jasplakinolide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, Jasplakinolide, ... | | Authors: | Hvorecny, K.L, Sladewski, T.E, Heaslip, A.T, Kollman, J.M. | | Deposit date: | 2023-08-09 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Toxoplasma gondii actin filaments are tuned for rapid disassembly and turnover.
Nat Commun, 15, 2024
|
|
8TRM
 
 | | Actin 1 from T. gondii in filaments bound to MgADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, MAGNESIUM ION | | Authors: | Hvorecny, K.L, Sladewski, T.E, Heaslip, A.T, Kollman, J.M. | | Deposit date: | 2023-08-09 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Toxoplasma gondii actin filaments are tuned for rapid disassembly and turnover.
Nat Commun, 15, 2024
|
|
8U8J
 
 | |
6PQ4
 
 | | LCP-embedded Proteinase K treated with lipase | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Bu, G, Zhu, L, Jing, L, Shi, D, Gonen, T, Liu, W, Nannenga, B.L. | | Deposit date: | 2019-07-08 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | Structure Determination from Lipidic Cubic Phase Embedded Microcrystals by MicroED.
Structure, 28, 2020
|
|
3KZA
 
 | | Crystal structure of Gyuba, a patched chimera of b-lactglobulin | | Descriptor: | Beta-lactoglobulin | | Authors: | Tsuge, H, Ohtomo, H, Utsunomiya, H, Konuma, T, Ikeguchi, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-12-22 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and stability of Gyuba, a patched chimera of b-lactoglobulin
Protein Sci., 20, 2011
|
|
3KYR
 
 | | Bace-1 in complex with a norstatine type inhibitor | | Descriptor: | 3-[[(2S)-2-[[[(2S)-2-[[(2S)-2-[[(2S)-2-azanyl-3-(1H-1,2,3,4-tetrazol-5-ylcarbonylamino)propanoyl]amino]-3-methyl-butanoyl]amino]-4-methyl-pentanoyl]amino]methyl]-2-hydroxy-4-phenyl-butanoyl]amino]benzoic acid, Beta-secretase 1 | | Authors: | Lindberg, J.D, Borkakoti, N, Derbyshire, D, Nystrom, S. | | Deposit date: | 2009-12-07 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigation of a-phenylnorstatine and a-benzylnorstatine as transition state isostere motifs in the search for new BACE-1 inhibiotrs
To be Published
|
|
3KZJ
 
 | | Structure of complement Factor H variant R1203A | | Descriptor: | Complement factor H, SULFATE ION | | Authors: | Bhattacharjee, A, Lehtinen, M.J, Kajander, T, Goldman, A, Jokiranta, T.S. | | Deposit date: | 2009-12-08 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Both domain 19 and domain 20 of factor H are involved in binding to complement C3b and C3d
Mol.Immunol., 47, 2010
|
|
8U1S
 
 | |
4Y00
 
 | | Crystal Structure of Human TDP-43 RRM1 Domain with D169G Mutation in Complex with an Unmodified Single-stranded DNA | | Descriptor: | DNA (5'-D(P*TP*TP*GP*AP*GP*CP*GP*T)-3'), TAR DNA-binding protein 43 | | Authors: | Chiang, C.H, Kuo, P.H, Yang, W.Z, Yuan, H.S. | | Deposit date: | 2015-02-05 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of disease-related TDP-43 D169G mutation: linking enhanced stability and caspase cleavage efficiency to protein accumulation
Sci Rep, 6, 2016
|
|
8U1Q
 
 | |
8U8K
 
 | |
3L59
 
 | | Structure of BACE Bound to SCH710413 | | Descriptor: | (2Z)-3-(3-chlorobenzyl)-2-imino-5,5-dimethylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3KN5
 
 | |
3KM2
 
 | |
