6XIG
 
 | |
2A5H
 
 | | 2.1 Angstrom X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale SB4, with Michaelis analog (L-alpha-lysine external aldimine form of pyridoxal-5'-phosphate). | | Descriptor: | IRON/SULFUR CLUSTER, L-lysine 2,3-aminomutase, LYSINE, ... | | Authors: | Lepore, B.W, Ruzicka, F.J, Frey, P.A, Ringe, D. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1OLT
 
 | | Coproporphyrinogen III oxidase (HemN) from Escherichia coli is a Radical SAM enzyme. | | Descriptor: | IRON/SULFUR CLUSTER, OXYGEN-INDEPENDENT COPROPORPHYRINOGEN III OXIDASE, S-ADENOSYLMETHIONINE | | Authors: | Layer, G, Moser, J, Heinz, D.W, Jahn, D, Schubert, W.-D. | | Deposit date: | 2003-08-13 | | Release date: | 2003-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of Coproporphyrinogen III Oxidase Reveals Cofactor Geometry of Radical Sam Enzymes
Embo J., 22, 2003
|
|
2QGQ
 
 | | Crystal structure of TM_1862 from Thermotoga maritima. Northeast Structural Genomics Consortium target VR77 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Protein TM_1862 | | Authors: | Forouhar, F, Neely, H, Hussain, M, Seetharaman, J, Fang, Y, Chen, C.X, Cunningham, K, Conover, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Post-translational Modification of Ribosomal Proteins: STRUCTURAL AND FUNCTIONAL CHARACTERIZATION OF RimO FROM THERMOTOGA MARITIMA, A RADICAL S-ADENOSYLMETHIONINE METHYLTHIOTRANSFERASE.
J.Biol.Chem., 285, 2010
|
|
6XI9
 
 | | X-ray crystal structure of MqnE from Pedobacter heparinus in complex with aminofutalosine and methionine | | Descriptor: | 9-[7-(3-carboxyphenyl)-5,6-dideoxy-beta-D-ribo-heptodialdo-1,4-furanosyl]-9H-purin-6-amine, Aminodeoxyfutalosine synthase, CHLORIDE ION, ... | | Authors: | Grove, T.L, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Narrow-Spectrum Antibiotic Targeting of the Radical SAM Enzyme MqnE in Menaquinone Biosynthesis.
Biochemistry, 59, 2020
|
|
3C8F
 
 | |
6B4C
 
 | | Structure of Viperin from Trichoderma virens | | Descriptor: | CITRATE ANION, SULFATE ION, Viperin | | Authors: | Huang, R.H, Selvadurai, K. | | Deposit date: | 2017-09-26 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Reconstitution and substrate specificity for isopentenyl pyrophosphate of the antiviral radical SAM enzyme viperin.
J.Biol.Chem., 293, 2018
|
|
7PD1
 
 | | Crystal structure of the L-tyrosine-bound radical SAM tyrosine lyase ThiH (2-iminoacetate synthase) from Thermosinus carboxydivorans | | Descriptor: | 5'-DEOXYADENOSINE, BROMIDE ION, GLYCEROL, ... | | Authors: | Amara, P, Saragaglia, C, Mouesca, J.-M, Martin, L, Nicolet, Y. | | Deposit date: | 2021-08-04 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | L-tyrosine-bound ThiH structure reveals C-C bond break differences within radical SAM aromatic amino acid lyases.
Nat Commun, 13, 2022
|
|
7PD2
 
 | | Crystal structure of the substrate-free radical SAM tyrosine lyase ThiH (2-iminoacetate synthase) from Thermosinus carboxydivorans | | Descriptor: | 5'-DEOXYADENOSINE, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Amara, P, Saragaglia, C, Mouesca, J.-M, Martin, L, Nicolet, Y. | | Deposit date: | 2021-08-04 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | L-tyrosine-bound ThiH structure reveals C-C bond break differences within radical SAM aromatic amino acid lyases.
Nat Commun, 13, 2022
|
|
5L7L
 
 | |
5L7J
 
 | |
6C8V
 
 | | X-ray structure of PqqE from Methylobacterium extorquens | | Descriptor: | Coenzyme PQQ synthesis protein E, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Gizzi, A.S, Grove, T.L, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray and EPR Characterization of the Auxiliary Fe-S Clusters in the Radical SAM Enzyme PqqE.
Biochemistry, 57, 2018
|
|
2FB3
 
 | | Structure of MoaA in complex with 5'-GTP | | Descriptor: | 5'-DEOXYADENOSINE, GUANOSINE-5'-TRIPHOSPHATE, IRON/SULFUR CLUSTER, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2005-12-08 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Binding of 5'-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2FB2
 
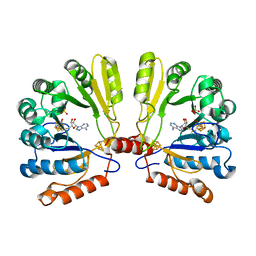 | | Structure of the MoaA Arg17/266/268/Ala triple mutant | | Descriptor: | IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, S-ADENOSYLMETHIONINE, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2005-12-08 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Binding of 5'-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
8VDW
 
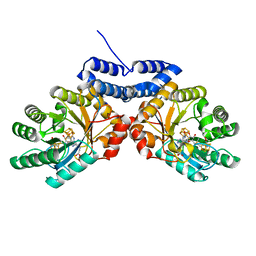 | |
8VCW
 
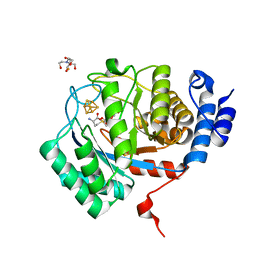 | | X-Ray Crystal Structure of the biotin synthase from B. obeum | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, Biotin synthase, ... | | Authors: | Lachowicz, J.C, Grove, T.L. | | Deposit date: | 2023-12-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of a Biotin Synthase That Utilizes an Auxiliary 4Fe-5S Cluster for Sulfur Insertion.
J.Am.Chem.Soc., 146, 2024
|
|
8VPO
 
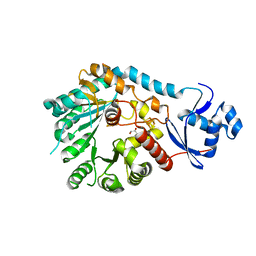 | | X-Ray Crystal Structure of TigE from Paramaledivibacter caminithermalis | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Radical SAM core domain-containing protein | | Authors: | Grove, T.L, Lachowicz, J.C, Zizola, C. | | Deposit date: | 2024-01-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural, Biochemical, and Bioinformatic Basis for Identifying Radical SAM Cyclopropyl Synthases.
Acs Chem.Biol., 19, 2024
|
|
6EFN
 
 | | Structure of a RiPP maturase, SkfB | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Grell, T.A.J, Drennan, C.L. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.291 Å) | | Cite: | Structure of a RiPP maturase, SkfB
J.Biol.Chem., 2018
|
|
4RTB
 
 | | X-ray structure of the FeFe-hydrogenase maturase HydG from Carboxydothermus hydrogenoformans | | Descriptor: | CHLORIDE ION, HydG protein, IRON/SULFUR CLUSTER, ... | | Authors: | Nicolet, Y, Pagnier, A, Zeppieri, L, Martin, L, Amara, P, Fontecilla-Camps, J.C. | | Deposit date: | 2014-11-14 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structure of HydG from Carboxydothermus hydrogenoformans: A Trifunctional [FeFe]-Hydrogenase Maturase.
Chembiochem, 16, 2015
|
|
8FOL
 
 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to SAM, alternate crystal form | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | Authors: | Moody, J.D, Saxton, A.J, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2022-12-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
8FO0
 
 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to a partially cleaved SAM molecule | | Descriptor: | IRON/SULFUR CLUSTER, POTASSIUM ION, Pyruvate formate-lyase 1-activating enzyme, ... | | Authors: | Moody, J.D, Saxton, A.J, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2022-12-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
8FSI
 
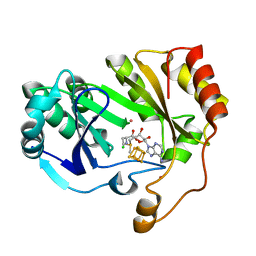 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to SAM | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | Authors: | Moody, J.D, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
2Z2U
 
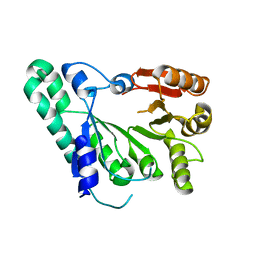 | | Crystal structure of archaeal TYW1 | | Descriptor: | UPF0026 protein MJ0257 | | Authors: | Suzuki, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2007-05-28 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Radical SAM Enzyme Catalyzing Tricyclic Modified Base Formation in tRNA
J.Mol.Biol., 372, 2007
|
|
1TV8
 
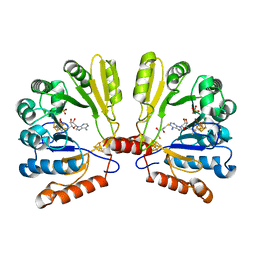 | | Structure of MoaA in complex with S-adenosylmethionine | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2004-06-28 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
3RF9
 
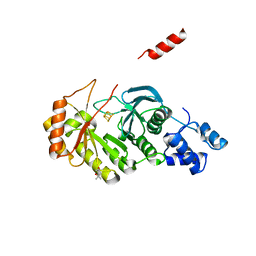 | | X-ray structure of RlmN from Escherichia coli | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, IRON/SULFUR CLUSTER, Ribosomal RNA large subunit methyltransferase N | | Authors: | Boal, A.K, Grove, T.L, McLaughlin, M.I, Yennawar, N, Booker, S.J, Rosenzweig, A.C. | | Deposit date: | 2011-04-05 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for methyl transfer by a radical SAM enzyme.
Science, 332, 2011
|
|
