7JQY
 
 | |
6GXT
 
 | | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography: FAcD2052MS after reaction initiation | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Mueller-Werkmeister, H, Tellkamp, F, Stuart, W, Persch, E, De Gasparo, R, Diederich, F, Pai, E.F, Miller, R.J.D. | | Deposit date: | 2018-06-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography.
Nat. Methods, 15, 2018
|
|
4OSE
 
 | |
4BAT
 
 | |
4B9E
 
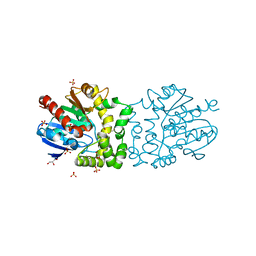 | | Structure of a putative epoxide hydrolase from Pseudomonas aeruginosa, with bound MFA. | | Descriptor: | GLYCEROL, PROBABLE EPOXIDE HYDROLASE, SULFATE ION, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-04 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1SCQ
 
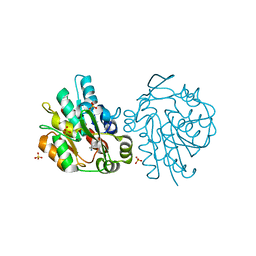 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetonecyanohydrin | | Descriptor: | (S)-acetone-cyanohydrin lyase, 2-HYDROXY-2-METHYLPROPANENITRILE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
4B9A
 
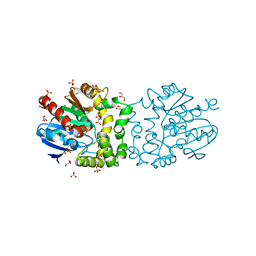 | | Structure of a putative epoxide hydrolase from Pseudomonas aeruginosa. | | Descriptor: | GLYCEROL, PROBABLE EPOXIDE HYDROLASE, SULFATE ION | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-03 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4BB0
 
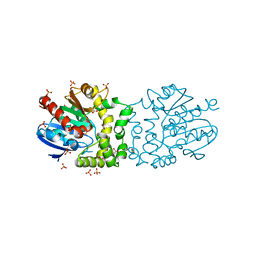 | |
4BAU
 
 | | Structure of a putative epoxide hydrolase t131d mutant from Pseudomonas aeruginosa, with bound MFA | | Descriptor: | CHLORIDE ION, PROBABLE EPOXIDE HYDROLASE, SULFATE ION, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-16 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a Putative Epoxide Hydrolase T131D Mutant from Pseudomonas Aeruginosa, with Bound Mfa
To be Published
|
|
1SC9
 
 | | Hydroxynitrile Lyase from Hevea brasiliensis in complex with the natural substrate acetone cyanohydrin | | Descriptor: | (S)-acetone-cyanohydrin lyase, 2-HYDROXY-2-METHYLPROPANENITRILE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
7ZJT
 
 | | Crystal structure of HsaD from Mycobacterium tuberculosis at 1.96 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, CHLORIDE ION | | Authors: | Barelier, S, Roig-Zamboni, V, Cavalier, J.F, Sulzenbacher, G. | | Deposit date: | 2022-04-11 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis.
Febs J., 290, 2023
|
|
7ZM2
 
 | | Crystal structure of HsaD from Mycobacterium tuberculosis in complex with Cyclophostin-like inhibitor CyC8b | | Descriptor: | 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, SULFATE ION, methoxy-[(3~{R})-3-[(2~{R})-1-methoxy-1,3-bis(oxidanylidene)butan-2-yl]pentadecyl]phosphinic acid | | Authors: | Barelier, S, Roig-Zamboni, V, Cavalier, J.F, Sulzenbacher, G. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis.
Febs J., 290, 2023
|
|
7ZM3
 
 | | Crystal structure of HsaD from Mycobacterium tuberculosis in complex with Cyclipostin-like inhibitor CyC17 | | Descriptor: | 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, SULFATE ION, hexadecyl dihydrogen phosphate | | Authors: | Barelier, S, Roig-Zamboni, V, Cavalier, J.F, Sulzenbacher, G. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis.
Febs J., 290, 2023
|
|
7ZM4
 
 | | Crystal structure of HsaD from Mycobacterium tuberculosis in complex with Cyclipostin-like inhibitor CyC31 | | Descriptor: | 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, SULFATE ION, undecyl dihydrogen phosphate | | Authors: | Barelier, S, Roig-Zamboni, V, Cavalier, J.F, Sulzenbacher, G. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis.
Febs J., 290, 2023
|
|
7ZM1
 
 | | Crystal structure of HsaD from Mycobacterium tuberculosis in complex with Cyclophostin-like inhibitor CyC7b | | Descriptor: | 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, SULFATE ION, methoxy-[(~{E},3~{R})-3-[(2~{R})-1-methoxy-1,3-bis(oxidanylidene)butan-2-yl]tridec-11-enyl]phosphinous acid | | Authors: | Barelier, S, Roig-Zamboni, V, Cavalier, J.F, Sulzenbacher, G. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis.
Febs J., 290, 2023
|
|
8SNI
 
 | | Hydroxynitrile Lyase from Hevea brasiliensis with Forty Mutations | | Descriptor: | (S)-hydroxynitrile lyase, 1,2-ETHANEDIOL, BENZOIC ACID, ... | | Authors: | Walsh, M.E, Greenberg, L.R, Kazlauskas, R.J, Pierce, C.T, Aihara, H, Evans, R.L, Shi, K. | | Deposit date: | 2023-04-27 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | To be published
To Be Published
|
|
8T87
 
 | |
1SCI
 
 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis | | Descriptor: | (S)-acetone-cyanohydrin lyase, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
8T88
 
 | |
1SCK
 
 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetone | | Descriptor: | (S)-acetone-cyanohydrin lyase, ACETONE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
4Q3L
 
 | | Crystal structure of MGS-M2, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | GLYCEROL, MGS-M2 | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
8SW8
 
 | | Crystal Structure of HaloTag7 bound to JF669-HaloTag ligand | | Descriptor: | 1-[(4aM,10P)-7-(azetidin-1-yl)-10-[2-carboxy-3,4,6-trifluoro-5-({2-[2-(hexyloxy)ethoxy]ethyl}carbamoyl)phenyl]-5,5-dimethyldibenzo[b,e]silin-3(5H)-ylidene]azetidin-1-ium, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Farrants, H, Schreiter, E.R. | | Deposit date: | 2023-05-17 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A modular chemigenetic calcium indicator for multiplexed in vivo functional imaging.
Nat.Methods, 2024
|
|
3NWO
 
 | |
5FLK
 
 | |
5EGN
 
 | |
