8G49
 
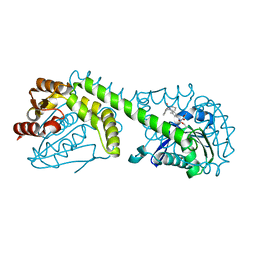 | | FphE, Staphylococcus aureus fluorophosphonate-binding serine hydrolases E, Oxadiazolone compound 3 bound | | Descriptor: | Fluorophosphonate-binding serine hydrolase E, methyl 2-formyl-2-[3-methyl-4-(3-phenoxybenzamido)phenyl]hydrazine-1-carboxylate | | Authors: | Fellner, M, Bakker, A.T, Martin, N.I, Stelt, M. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | FphE, Staphylococcus aureus fluorophosphonate-binding serine hydrolases E, Oxadiazolone compound 3 bound
To be published
|
|
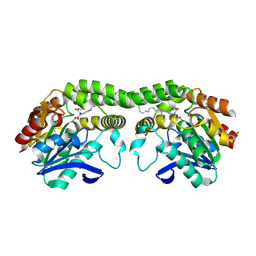
8T88
 
 | |
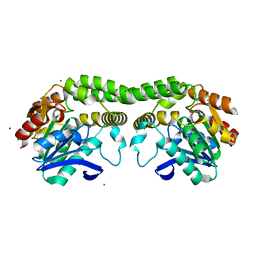
8T87
 
 | |
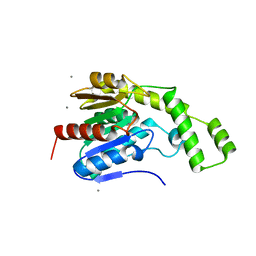
8FTP
 
 | |
8G0N
 
 | |
8G48
 
 | |
8SBQ
 
 | |
8S9J
 
 | |
8S9R
 
 | | SAL2, Staphylococcus aureus lipase 2 (geh, lip2), apo form | | Descriptor: | CALCIUM ION, Lipase 2, PHOSPHATE ION, ... | | Authors: | Fellner, M. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | SAL2, Staphylococcus aureus lipase 2 (geh, lip2), apo form
To Be Published
|
|
6OC5
 
 | | Lanthanide-dependent methanol dehydrogenase XoxF from Methylobacterium extorquens, in complex with Lanthanum | | Descriptor: | LANTHANUM (III) ION, Lanthanide-dependent methanol dehydrogenase XoxF | | Authors: | Fellner, M, Good, N.M, Martinez-Gomez, N.C, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-03-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lanthanide-dependent alcohol dehydrogenases require an essential aspartate residue for metal coordination and enzymatic function.
J.Biol.Chem., 295, 2020
|
|
6OC6
 
 | | Lanthanide-dependent methanol dehydrogenase XoxF from Methylobacterium extorquens, in complex with Lanthanum and Pyrroloquinoline quinone | | Descriptor: | LANTHANUM (III) ION, Lanthanide-dependent methanol dehydrogenase XoxF, PYRROLOQUINOLINE QUINONE | | Authors: | Fellner, M, Good, N.M, Martinez-Gomez, N.C, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-03-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Lanthanide-dependent alcohol dehydrogenases require an essential aspartate residue for metal coordination and enzymatic function.
J.Biol.Chem., 295, 2020
|
|
6D6Z
 
 | |
4TLF
 
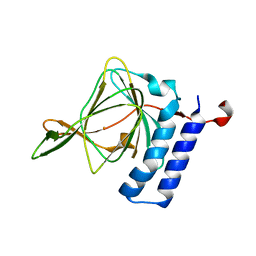 | | Crystal structure of Thiol dioxygenase from Pseudomonas aeruginosa | | Descriptor: | 3-mercaptopropionate dioxygenase, FE (II) ION | | Authors: | Fellner, M, Tchesnokov, E.P, Jameson, G.N.L, Wilbanks, S.M. | | Deposit date: | 2014-05-29 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | The Cysteine Dioxygenase Homologue from Pseudomonas aeruginosa Is a 3-Mercaptopropionate Dioxygenase.
J.Biol.Chem., 290, 2015
|
|
4UBG
 
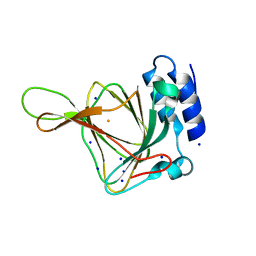 | | Resting state of rat cysteine dioxygenase C93G variant | | Descriptor: | Cysteine dioxygenase type 1, FE (II) ION, SODIUM ION | | Authors: | Fellner, M, Tchesnokov, E.P, Jameson, G.N, Wilbanks, S.M. | | Deposit date: | 2014-08-13 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Cys-Tyr Cross-Link of Cysteine Dioxygenase Changes the Optimal pH of the Reaction without a Structural Change.
Biochemistry, 53, 2014
|
|
4Z82
 
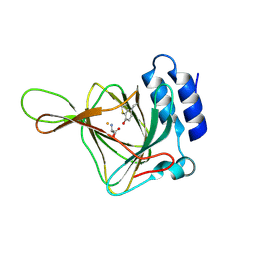 | | Cysteine bound rat cysteine dioxygenase C164S variant at pH 8.1 | | Descriptor: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION | | Authors: | Fellner, M, Tchesnokov, E.P, Siakkou, E, Rutledge, M.T, Kanitz, M, Jameson, G.N.L, Wilbanks, S.M. | | Deposit date: | 2015-04-08 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influence of cysteine 164 on active site structure in rat cysteine dioxygenase.
J.Biol.Inorg.Chem., 21, 2016
|
|
6VH9
 
 | | FphF, Staphylococcus aureus fluorophosphonate-binding serine hydrolases F, apo form | | Descriptor: | Esterase family protein, SODIUM ION | | Authors: | Fellner, M, Jamieson, S.A, Brewster, J.L, Mace, P.D. | | Deposit date: | 2020-01-09 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural Basis for the Inhibitor and Substrate Specificity of the Unique Fph Serine Hydrolases of Staphylococcus aureus .
Acs Infect Dis., 6, 2020
|
|
6VHD
 
 | |
6VHE
 
 | |
6WCX
 
 | |
6DG3
 
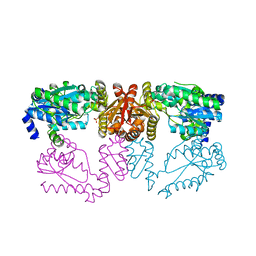 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with caesium | | Descriptor: | CESIUM ION, PHOSPHATE ION, Pyridinium-3,5-biscarboxylic acid mononucleotide sulfurtransferase | | Authors: | Fellner, M, Hausinger, R.P, Hu, J. | | Deposit date: | 2018-05-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.944 Å) | | Cite: | Unique cesium-binding sites in proteins, a case study with the sacrificial sulfur transferase LarE
J Life Sci, 2021
|
|
6ECJ
 
 | |
4WVZ
 
 | | Crystal structure of artificial crosslinked thiol dioxygenase G95C variant from Pseudomonas aeruginosa | | Descriptor: | 3-mercaptopropionate dioxygenase, FE (II) ION | | Authors: | Fellner, M, Tchesnokov, E.P, Jameson, G.N.L, Wilbanks, S.M. | | Deposit date: | 2014-11-09 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Substrate and pH-Dependent Kinetic Profile of 3-Mercaptopropionate Dioxygenase from Pseudomonas aeruginosa.
Biochemistry, 55, 2016
|
|
4YNI
 
 | |
4YSF
 
 | |
4YYO
 
 | | Resting state of rat cysteine dioxygenase C164S variant | | Descriptor: | CHLORIDE ION, Cysteine dioxygenase type 1, FE (II) ION | | Authors: | Fellner, M, Tchesnokov, E.P, Siakkou, E, Rutledge, M.T, Jameson, G.N.L, Wilbanks, S.M. | | Deposit date: | 2015-03-24 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Influence of cysteine 164 on active site structure in rat cysteine dioxygenase.
J.Biol.Inorg.Chem., 21, 2016
|
|
