8Y6Q
 
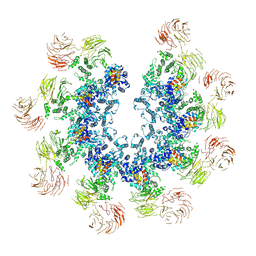 | | Structure of the Dark/Dronc complex | | Descriptor: | Apaf-1 related killer DARK, Caspase Dronc | | Authors: | Tian, L, Li, Y, Shi, Y. | | Deposit date: | 2024-02-03 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Dark and Dronc activation in Drosophila melanogaster.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8Y6P
 
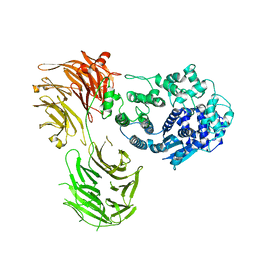 | |
8Y6I
 
 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in nanodisc | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ATP-dependent translocase ABCB1,mNeonGreen, CHOLESTEROL, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
8Y6H
 
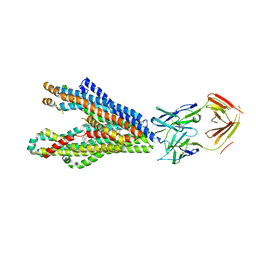 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in LMNG detergent | | Descriptor: | ATP-dependent translocase ABCB1,mNeonGreen, UIC2 Fab heavy chain, UIC2 Fab light chain, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
8Y6F
 
 | |
8Y6D
 
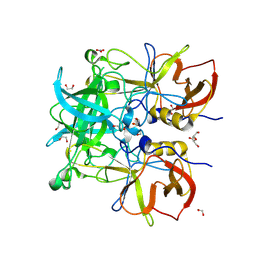 | | Norovirus GII.10 P domain and 2'-FL (tablet) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GII.10 norovirus P domain in complex with 2'-FL (tablet), ... | | Authors: | Hansman, G, Tame, J.R.H, Kher, G, Pancera, M. | | Deposit date: | 2024-02-02 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Norovirus GII.10 P domain and 2'-FL (tablet)
To Be Published
|
|
8Y6C
 
 | | Norovirus GII.10 P domain and 2'-FL (powder) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GII.10 norovirus P domain in complex with 2'FL (powder), ... | | Authors: | Hansman, G, Tame, J.R.H, Kher, G, Pancera, M. | | Deposit date: | 2024-02-02 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Norovirus GII.10 P domain and 2'-FL (powder)
To Be Published
|
|
8Y66
 
 | | Cryo-EM structure of human urate transporter GLUT9 bound to inhibitor apigenin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Solute carrier family 2, facilitated glucose transporter member 9 | | Authors: | Pan, X.J, Shen, Z.L, Xu, L, Huang, G.X.Y. | | Deposit date: | 2024-02-01 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for urate recognition and apigenin inhibition of human GLUT9.
Nat Commun, 15, 2024
|
|
8Y65
 
 | | Cryo-EM structure of human urate transporter GLUT9 bound to substrate urate | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 9, URIC ACID | | Authors: | Pan, X.J, Shen, Z.L, Xu, L, Huang, G.X.Y. | | Deposit date: | 2024-02-01 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural basis for urate recognition and apigenin inhibition of human GLUT9.
Nat Commun, 15, 2024
|
|
8Y5V
 
 | | GII.4 Sydney PD and 2'-FL | | Descriptor: | 1,2-ETHANEDIOL, GII.4 Sydney PD in complex with 2'-FL (powder), alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hansman, G, Tame, J.R.H, Kher, G, Pancera, M. | | Deposit date: | 2024-02-01 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | GII.4 Sydney PD and 2'-FL
To Be Published
|
|
8Y53
 
 | | Cryo-EM structure of the MK-5046-bound BRS3-Gq complex | | Descriptor: | (2~{S})-1,1,1-tris(fluoranyl)-2-(4-pyrazol-1-ylphenyl)-3-[5-[[1-(trifluoromethyl)cyclopropyl]methyl]-1~{H}-imidazol-2-yl]propan-2-ol, Bombesin receptor subtype-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, C, Xu, Y, Yin, W, Xu, H.E. | | Deposit date: | 2024-01-31 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural insights into ligand recognition, selectivity, and activation of bombesin receptor subtype-3.
Cell Rep, 43, 2024
|
|
8Y52
 
 | | Cryo-EM structure of the BA1-bound BRS3-Gq complex | | Descriptor: | BA1, Bombesin receptor subtype-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, C, Xu, Y, Yin, W, Xu, H.E. | | Deposit date: | 2024-01-31 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into ligand recognition, selectivity, and activation of bombesin receptor subtype-3.
Cell Rep, 43, 2024
|
|
8Y4J
 
 | |
8Y46
 
 | | Crystal structure of L-2-keto-3-deoxyfuconate 4-dehydrogenase bound to L-KDF or L-2,4-DKDF | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, L-2,4-diketo-3-deoxyfuconate, ... | | Authors: | Akagashi, M, Watanabe, S. | | Deposit date: | 2024-01-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structure of L-2-keto-3-deoxyfuconate 4-dehydrogenase reveals a unique binding mode as a alpha-furanosyl hemiketal of substrates.
Sci Rep, 14, 2024
|
|
8Y3X
 
 | | Cell divisome sPG hydrolysis machinery FtsEX-EnvC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Zhang, Z, Dong, H, Chen, Y. | | Deposit date: | 2024-01-29 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structure and activity of the septal peptidoglycan hydrolysis machinery crucial for bacterial cell division.
Plos Biol., 22, 2024
|
|
8Y33
 
 | | A near-infrared fluorescent protein of de novo backbone design | | Descriptor: | 3-[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-2-[[5-[(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, near-infrared fluorescent protein | | Authors: | Hu, X, Xu, Y. | | Deposit date: | 2024-01-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Using Protein Design and Directed Evolution to Monomerize a Bright Near-Infrared Fluorescent Protein.
Acs Synth Biol, 13, 2024
|
|
8Y2S
 
 | | P-hydroxybenzoate hydroxylase complexed with 4-hydroxy-3-methylbenzoic acid | | Descriptor: | 3-methyl-4-oxidanyl-benzoic acid, 4-hydroxybenzoate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2024-01-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Enhancement of Flavin-Containing Monooxygenase through Machine Learning Methodology
Acs Catalysis, 14, 2024
|
|
8Y22
 
 | | FGFR1 kinase domain with a covalent inhibitor 9g | | Descriptor: | Fibroblast growth factor receptor 1, SULFATE ION, ~{N}-[4-[[4-azanyl-3-(7-methoxy-5-methyl-1-benzothiophen-2-yl)pyrazolo[3,4-d]pyrimidin-1-yl]methyl]phenyl]propanamide | | Authors: | Chen, X.J, Chen, Y.H. | | Deposit date: | 2024-01-25 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | Design, synthesis and biological evaluation of 5-amino-1H-pyrazole-4-carboxamide derivatives as pan-FGFR covalent inhibitors.
Eur.J.Med.Chem., 275, 2024
|
|
8Y1R
 
 | | in situ room temperature Laue crystallography | | Descriptor: | Lysozyme C | | Authors: | Wang, Z.J, Wang, S.S, Pan, Q.Y, Yu, L, Su, Z.H, Yang, T.Y, Wang, Y.Z, Zhang, W.Z, Hao, Q, Gao, X.Y. | | Deposit date: | 2024-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single pulse data collection with an X-ray chopper at in situ room temperature Laue crystallography beamline BL03HB
Nucl Instrum Methods Phys Res A, 1069, 2024
|
|
8Y1M
 
 | | Xylanase R from Bacillus sp. TAR-1 complexed with xylobiose. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Nakamura, T, Kuwata, K, Takita, T, Mizutani, K, Mikami, B, Nakamura, S, Yasukawa, K. | | Deposit date: | 2024-01-25 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR.
Sci Rep, 14, 2024
|
|
8Y1L
 
 | |
8Y14
 
 | | Beta-catenin Crystal Structure | | Descriptor: | Catenin beta-1 | | Authors: | Tim, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Beta-catenin Crystal Structure
To Be Published
|
|
8Y11
 
 | |
8Y0X
 
 | | Dormant ribosome with SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Implication of Stm1 in the protection of eIF5A, eEF2 and tRNA through dormant ribosomes.
Front Mol Biosci, 11, 2024
|
|
8Y0W
 
 | | dormant ribosome with eIF5A, eEF2 and SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Implication of Stm1 in the protection of eIF5A, eEF2 and tRNA through dormant ribosomes.
Front Mol Biosci, 11, 2024
|
|
